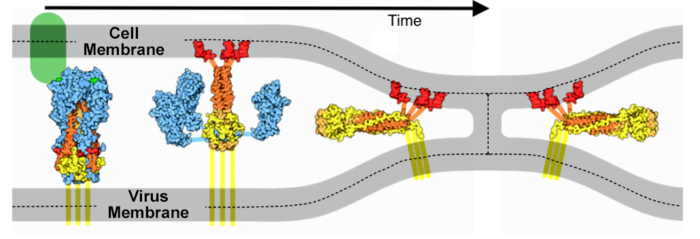
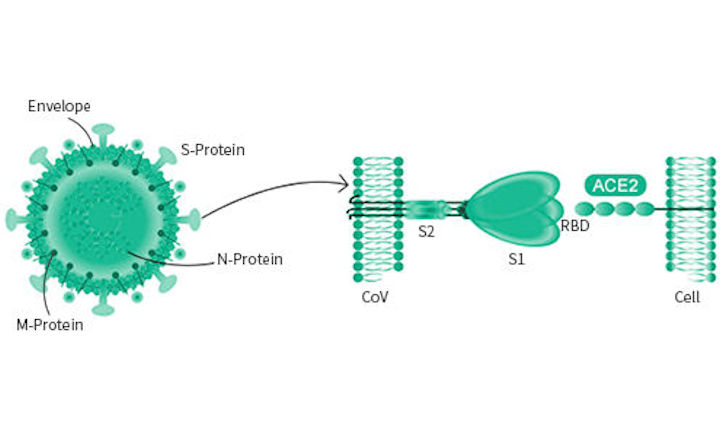
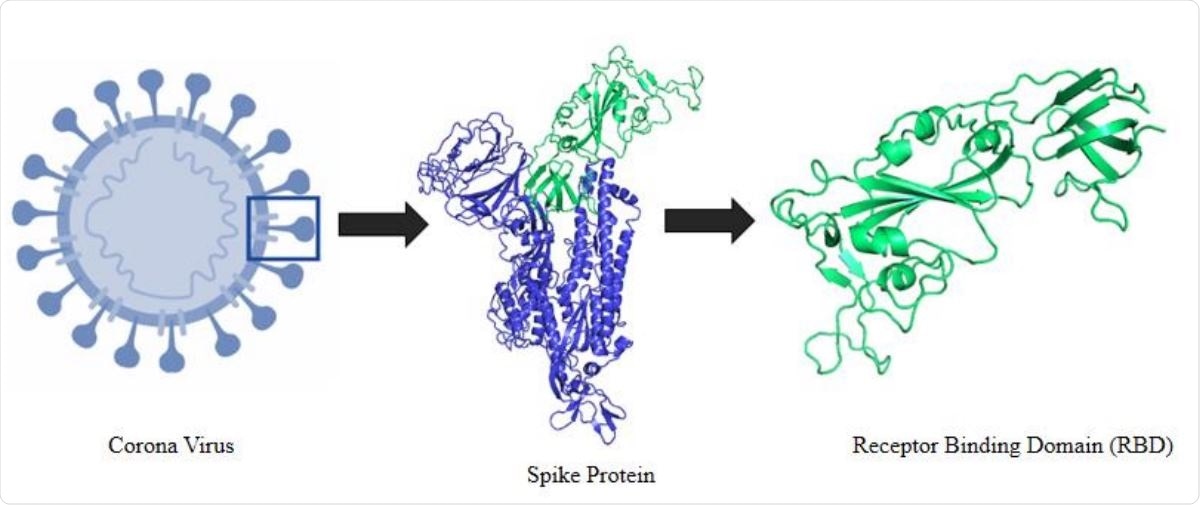
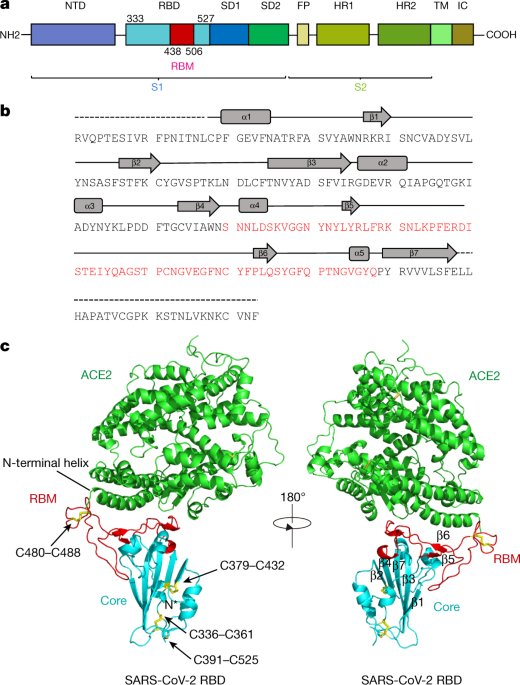
Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor | Nature Communications
![Dynamics of the interaction between the receptor-binding domain of SARS-CoV-2 Omicron (B.1.1.529) variant and human angiotensin-converting enzyme 2 [PeerJ] Dynamics of the interaction between the receptor-binding domain of SARS-CoV-2 Omicron (B.1.1.529) variant and human angiotensin-converting enzyme 2 [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/13680/1/fig-1-full.png)
Dynamics of the interaction between the receptor-binding domain of SARS-CoV-2 Omicron (B.1.1.529) variant and human angiotensin-converting enzyme 2 [PeerJ]
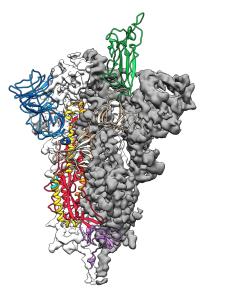
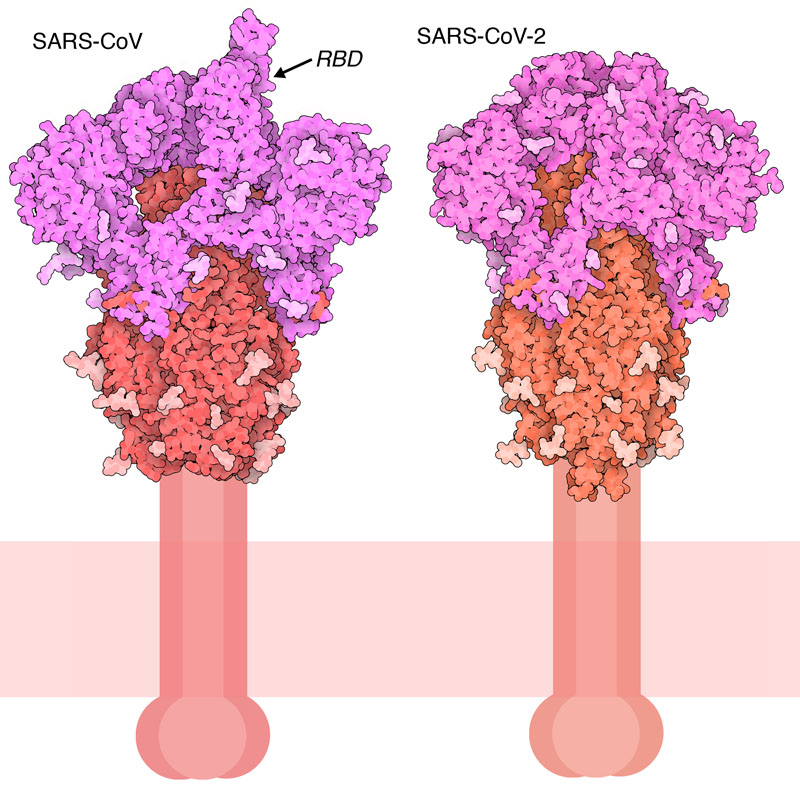
NIH Scientists Identify Atomic Structure of Novel Coronavirus Protein | NIAID: National Institute of Allergy and Infectious Diseases
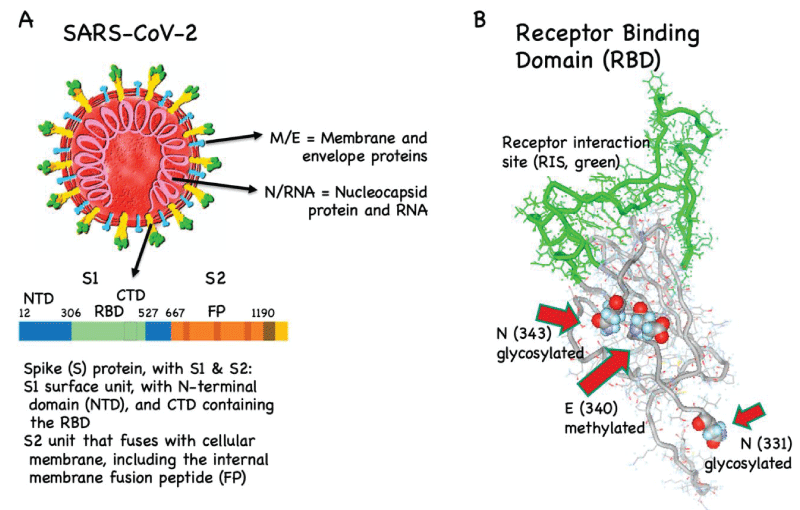
RBD targeted COVID vaccine and full length spike-protein vaccine (mutation and glycosylation role) relationship with procoagulant effect

The structural basis of accelerated host cell entry by SARS‐CoV‐2† - Seyran - 2021 - The FEBS Journal - Wiley Online Library
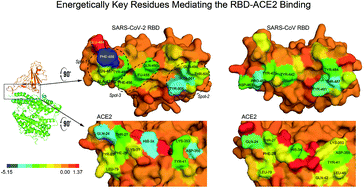
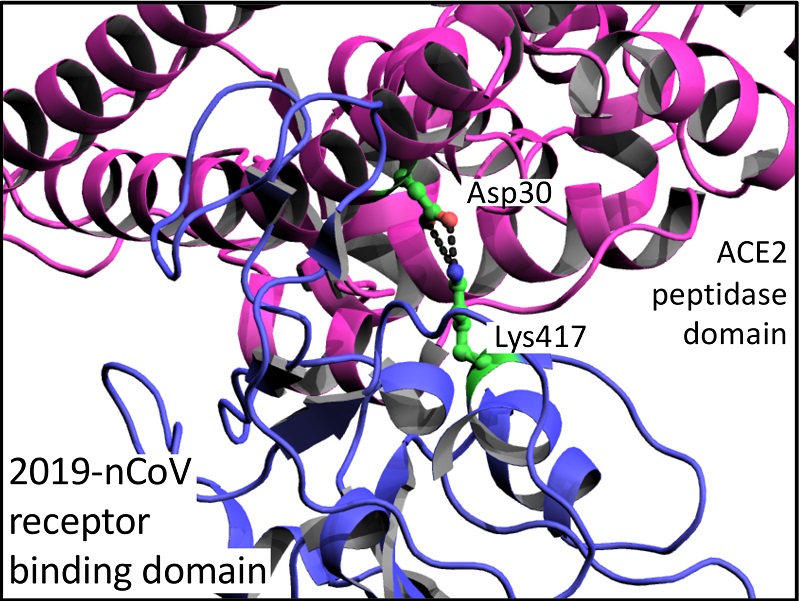
Key residues of the receptor binding domain in the spike protein of SARS-CoV-2 mediating the interactions with ACE2: a molecular dynamics study - Nanoscale (RSC Publishing)

SARS-CoV-2 IgG antibodies and why the receptor-binding domain of the spike protein is so important - YouTube

Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity - Computational and Structural Biotechnology Journal

Structural Characterization of N‐Linked Glycans in the Receptor Binding Domain of the SARS‐CoV‐2 Spike Protein and their Interactions with Human Lectins - Lenza - 2020 - Angewandte Chemie International Edition - Wiley Online Library
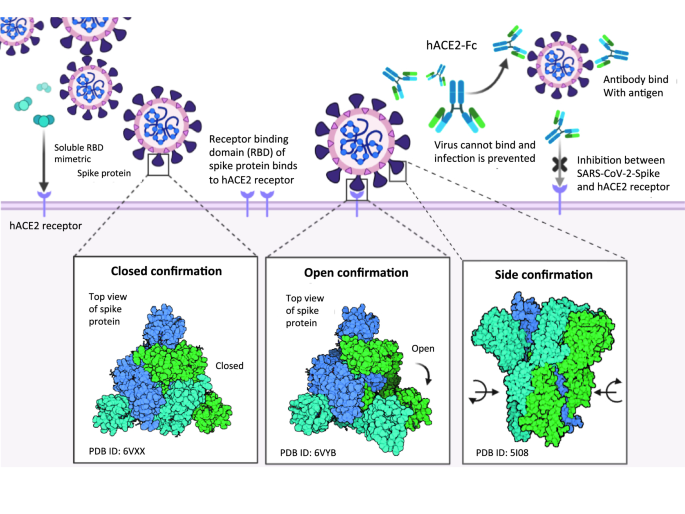
Monoclonal antibody designed for SARS-nCoV-2 spike protein of receptor binding domain on antigenic targeted epitopes for inhibition to prevent viral entry | Molecular Diversity

Targeting SARS-CoV-2 Receptor Binding Domain with Stapled Peptides: An In Silico Study | The Journal of Physical Chemistry B

Identification of a repurposed drug as an inhibitor of Spike protein of human coronavirus SARS-CoV-2 by computational methods | Journal of Biosciences

Computational screening of 645 antiviral peptides against the receptor-binding domain of the spike protein in SARS-CoV-2 - ScienceDirect

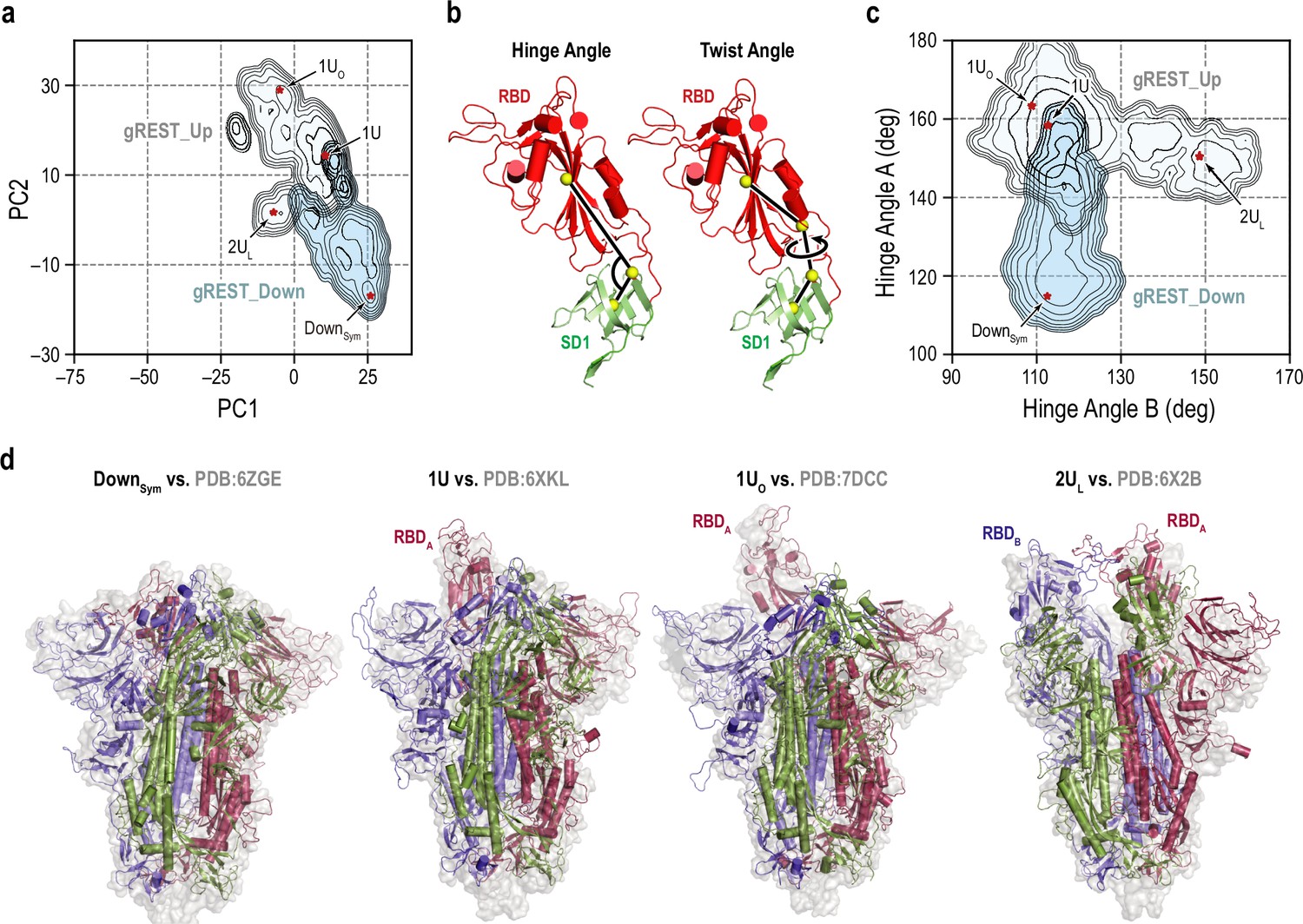
Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains - ScienceDirect

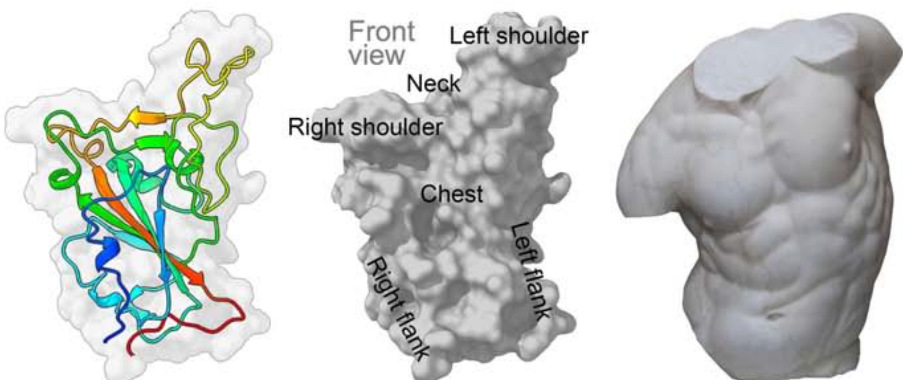
a) Structure of the receptor-binding domain (RBD) of the S protein in... | Download Scientific Diagram