RCSB PDB - 5XS8: Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with two sulfate groups at C-4 and C-6 positions of GalNAc

Solute transport systems found in bacteria and archaea. (A) Channels.... | Download Scientific Diagram

Structural studies of Mn-specific solute binding proteins (SBPs) from... | Download Scientific Diagram

The role of solute binding proteins in signal transduction - Computational and Structural Biotechnology Journal
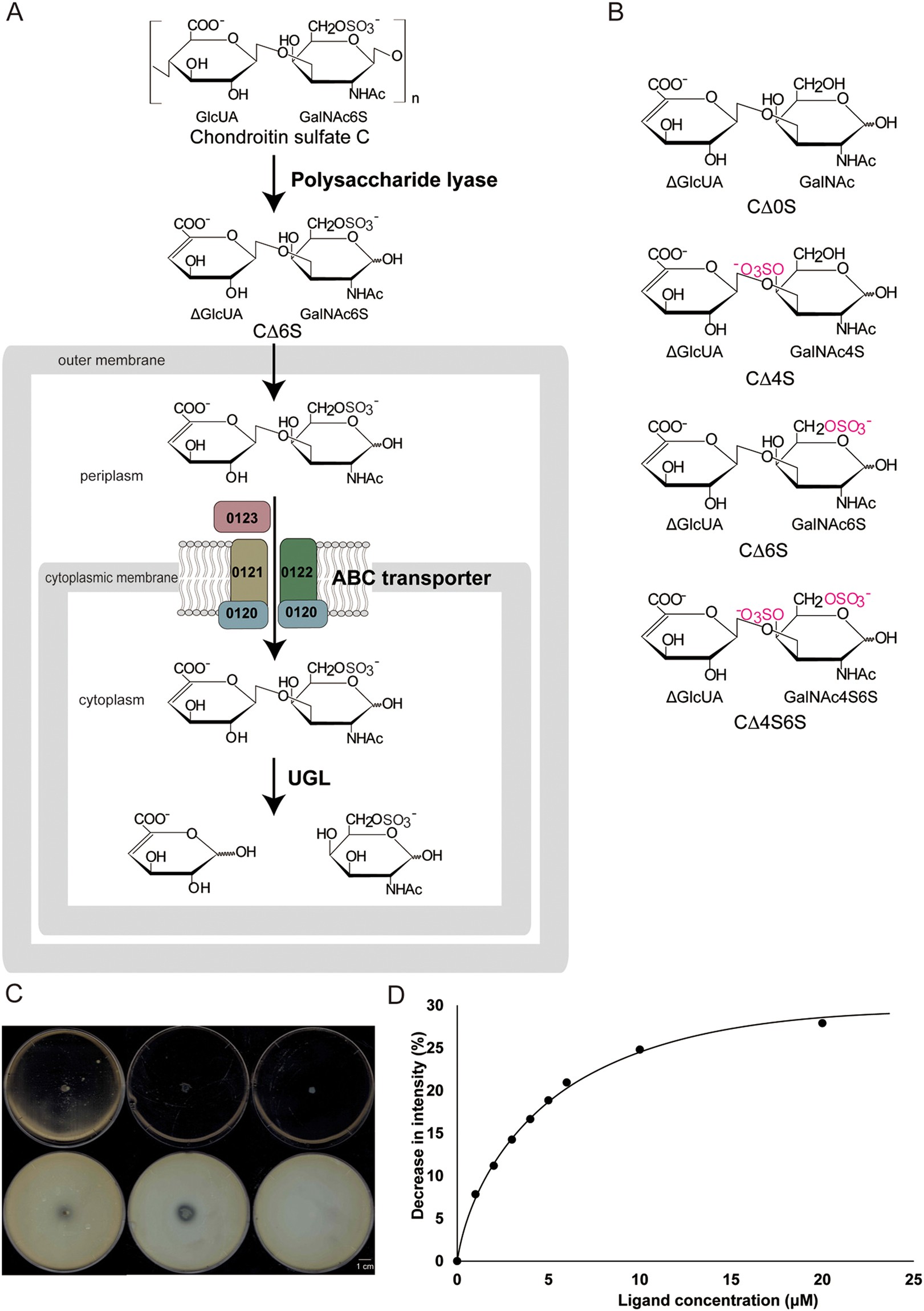
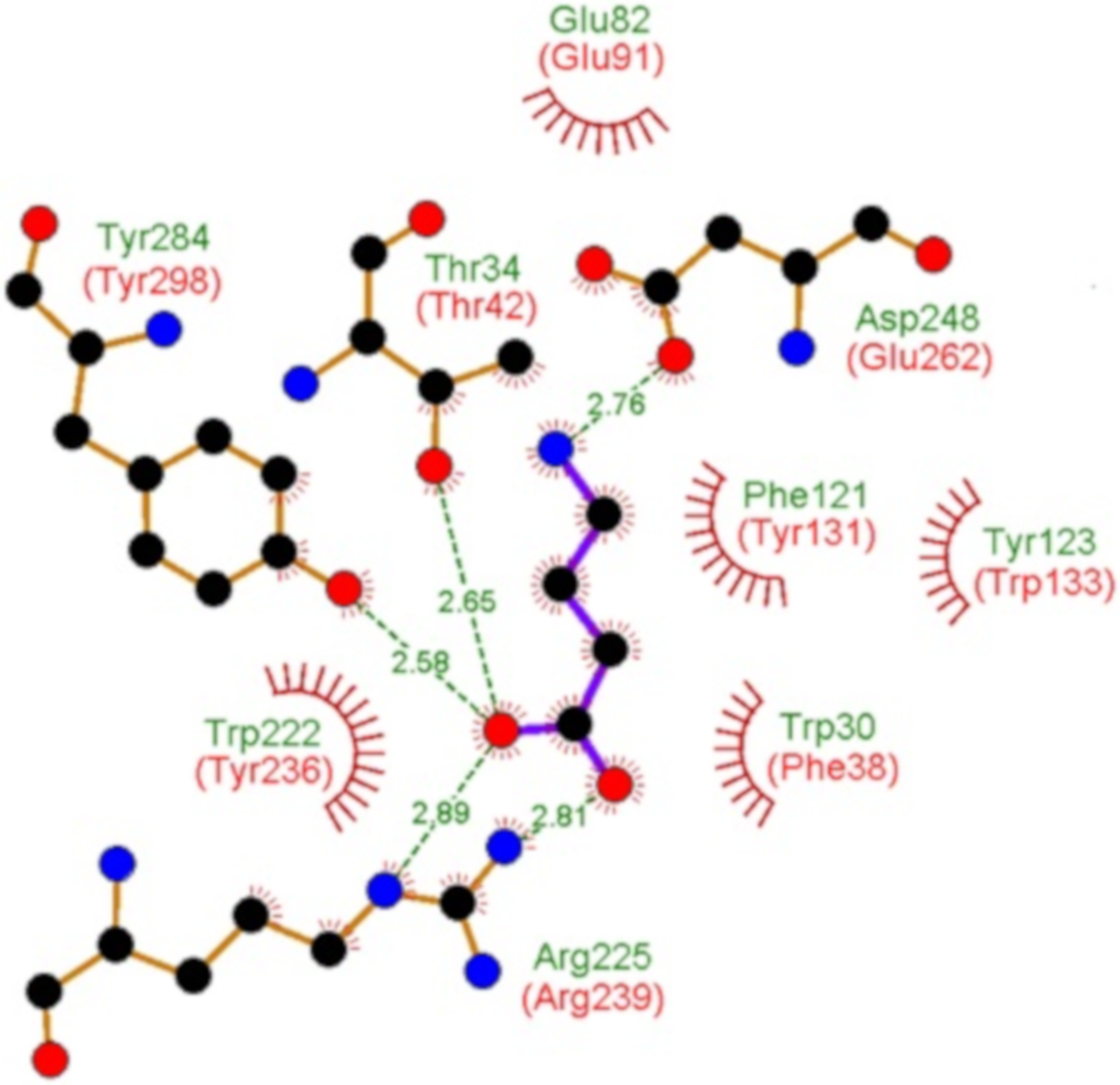
Alternative substrate-bound conformation of bacterial solute-binding protein involved in the import of mammalian host glycosaminoglycans | Scientific Reports
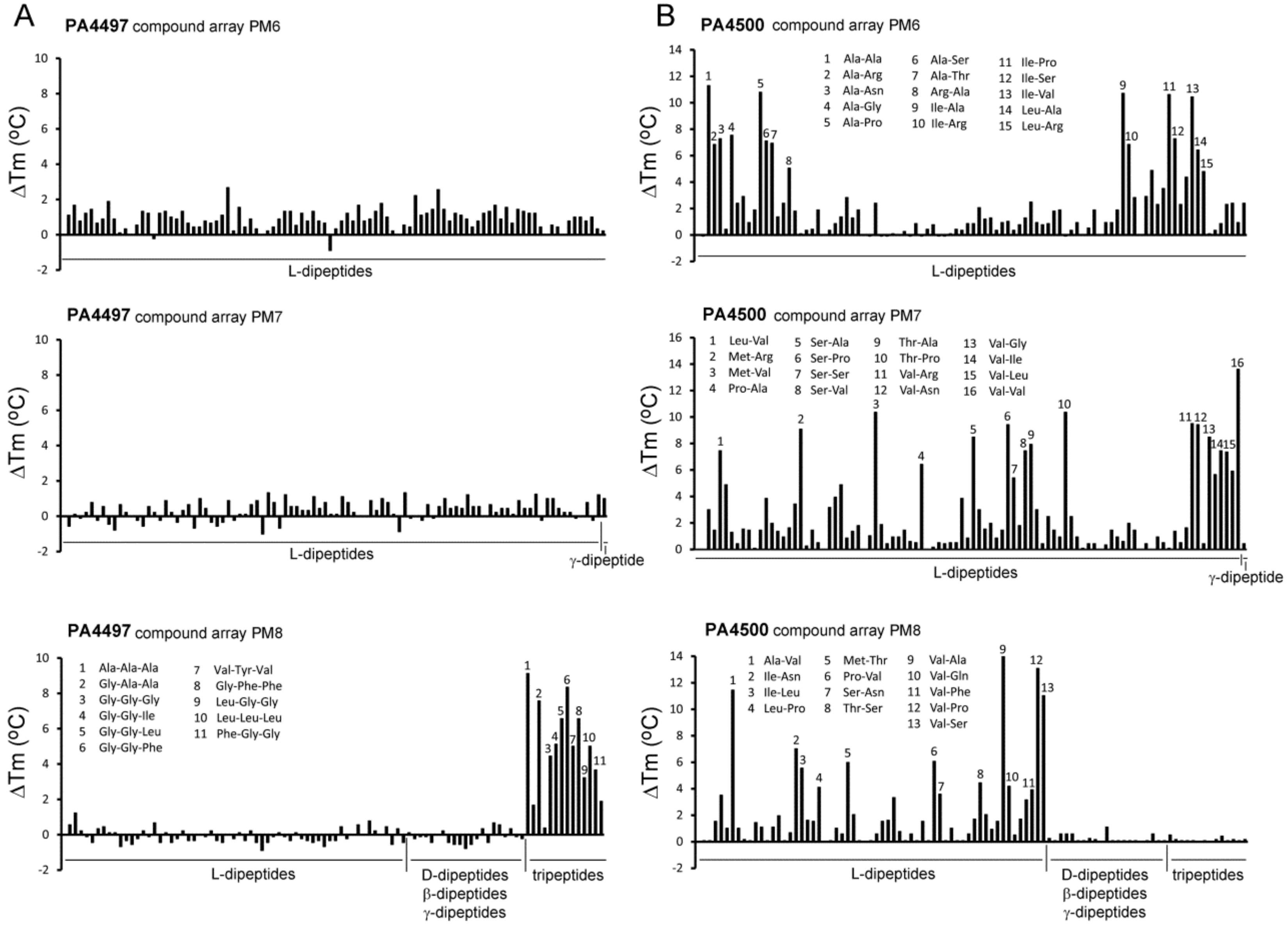
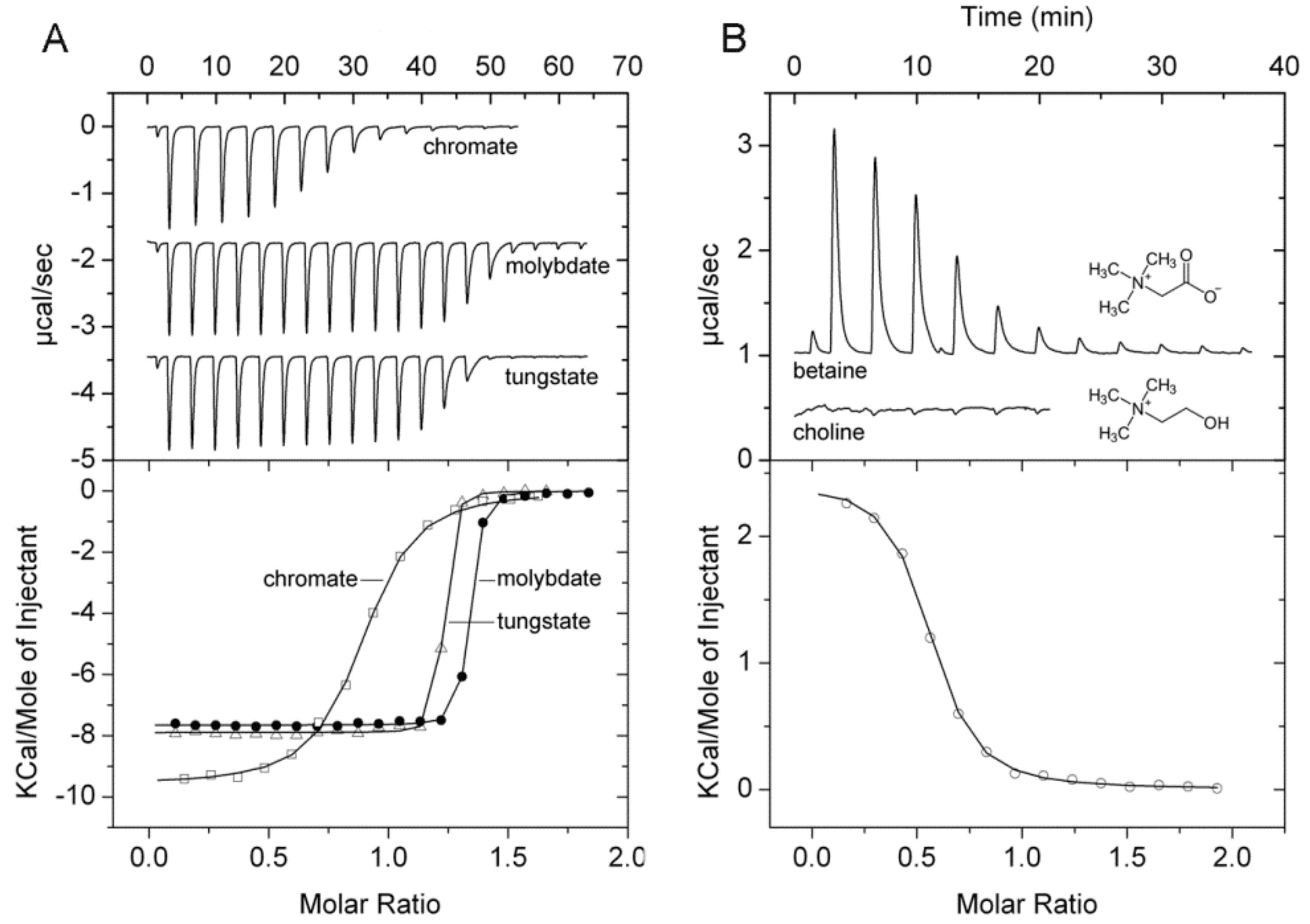
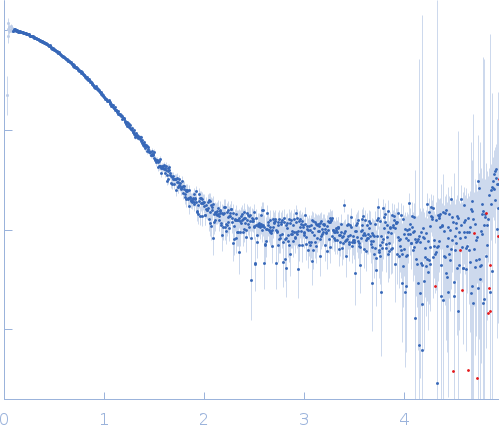
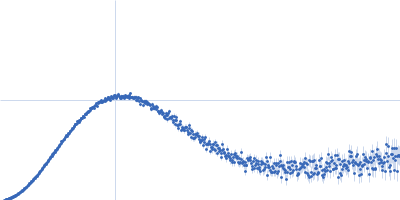
IJMS | Free Full-Text | Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins
IJMS | Free Full-Text | Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins
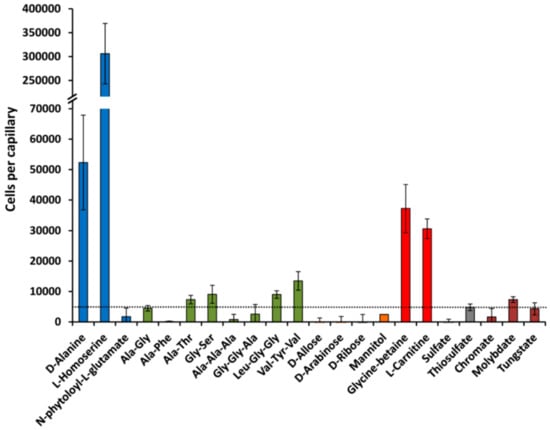
IJMS | Free Full-Text | Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins

Altered conformational sampling along an evolutionary trajectory changes the catalytic activity of an enzyme | Nature Communications

IJMS | Free Full-Text | Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins
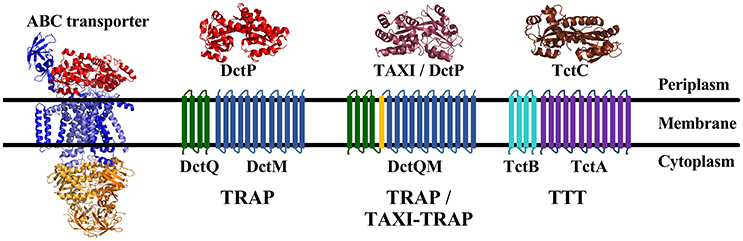
Frontiers | Tripartite ATP-Independent Periplasmic (TRAP) Transporters and Tripartite Tricarboxylate Transporters (TTT): From Uptake to Pathogenicity

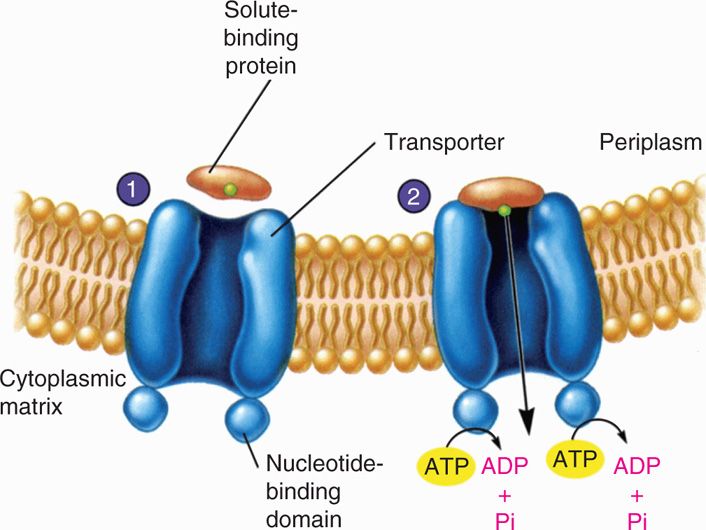
Overview and differences between the ATP-binding cassette (ABC) and... | Download Scientific Diagram

RCSB PDB - 4NX1: Crystal structure of a trap periplasmic solute binding protein from Sulfitobacter sp. nas-14.1, target EFI-510292, with bound alpha-D-taluronate

The role of solute binding proteins in signal transduction - Computational and Structural Biotechnology Journal
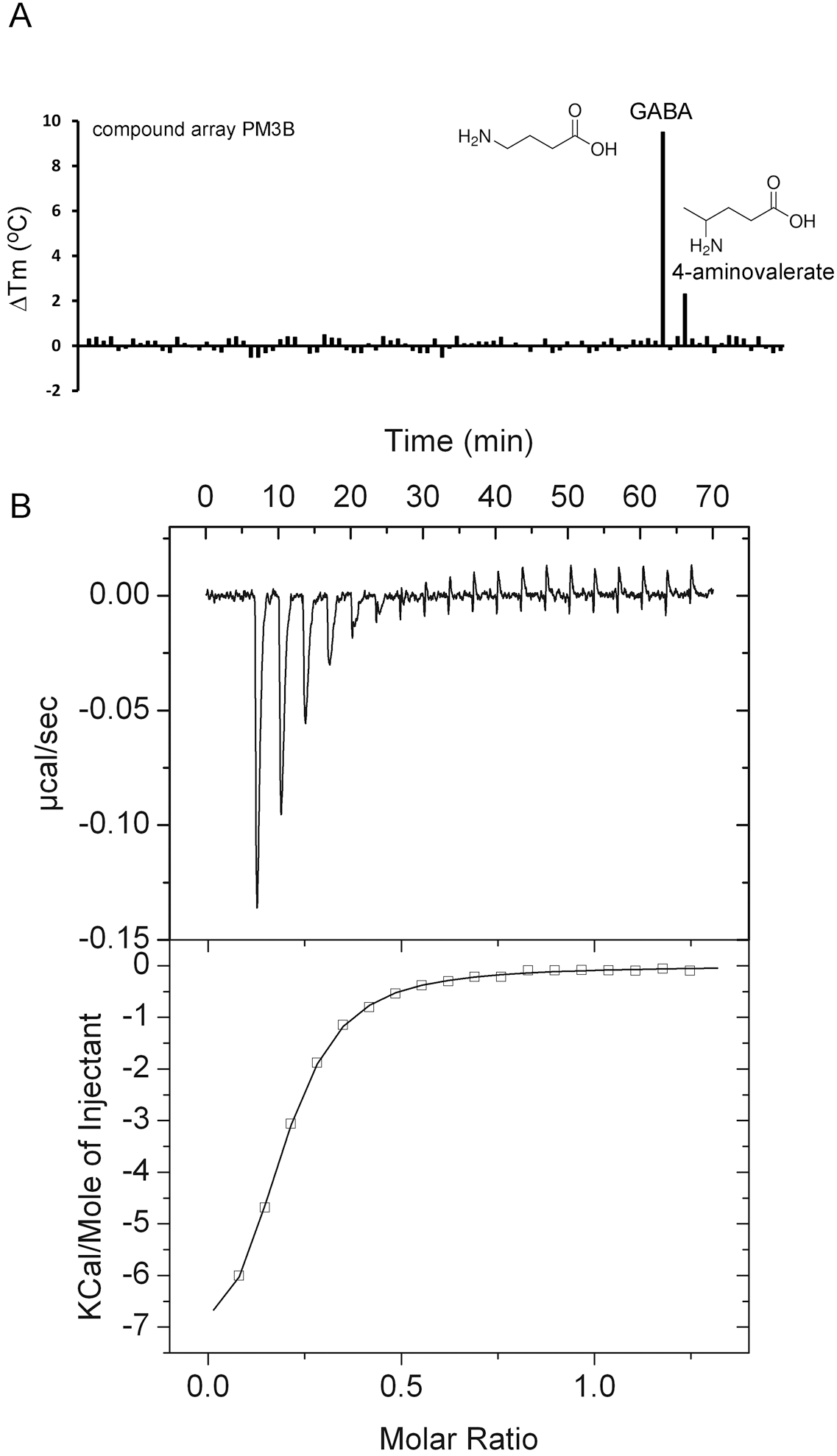
IJMS | Free Full-Text | Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins
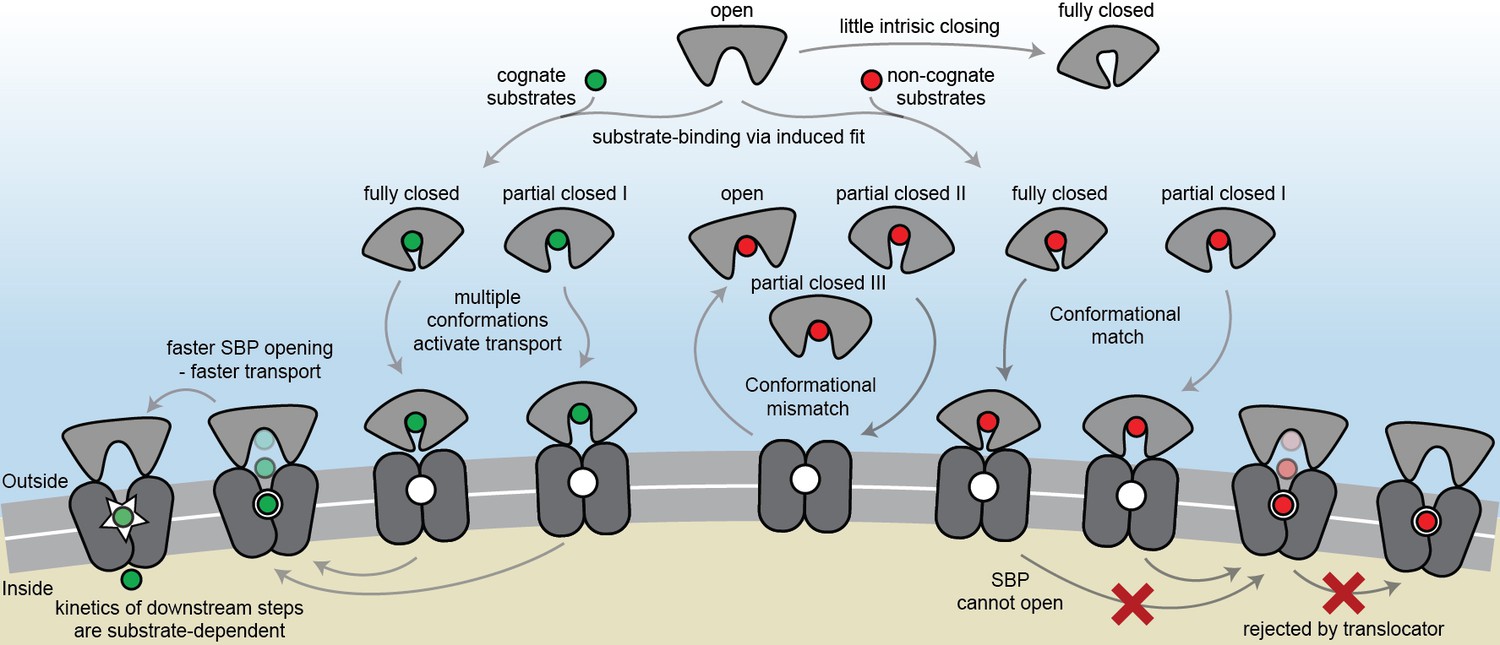
Conformational and dynamic plasticity in substrate-binding proteins underlies selective transport in ABC importers | eLife
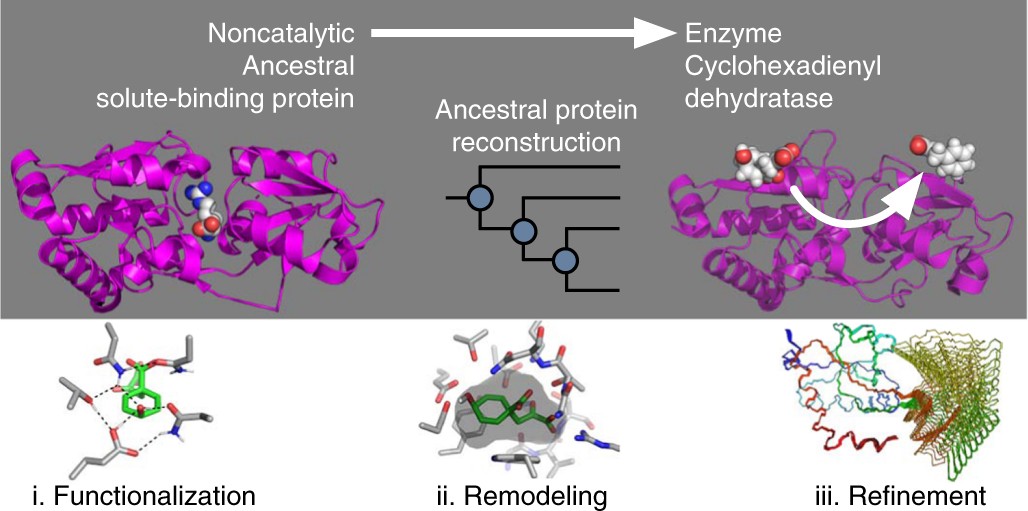
Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein | Nature Chemical Biology

A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate - ScienceDirect

RCSB PDB - 5XS8: Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with two sulfate groups at C-4 and C-6 positions of GalNAc