RNA-RNA binding prediction and interaction of hsa-miR-1-3p and ODC.... | Download Scientific Diagram
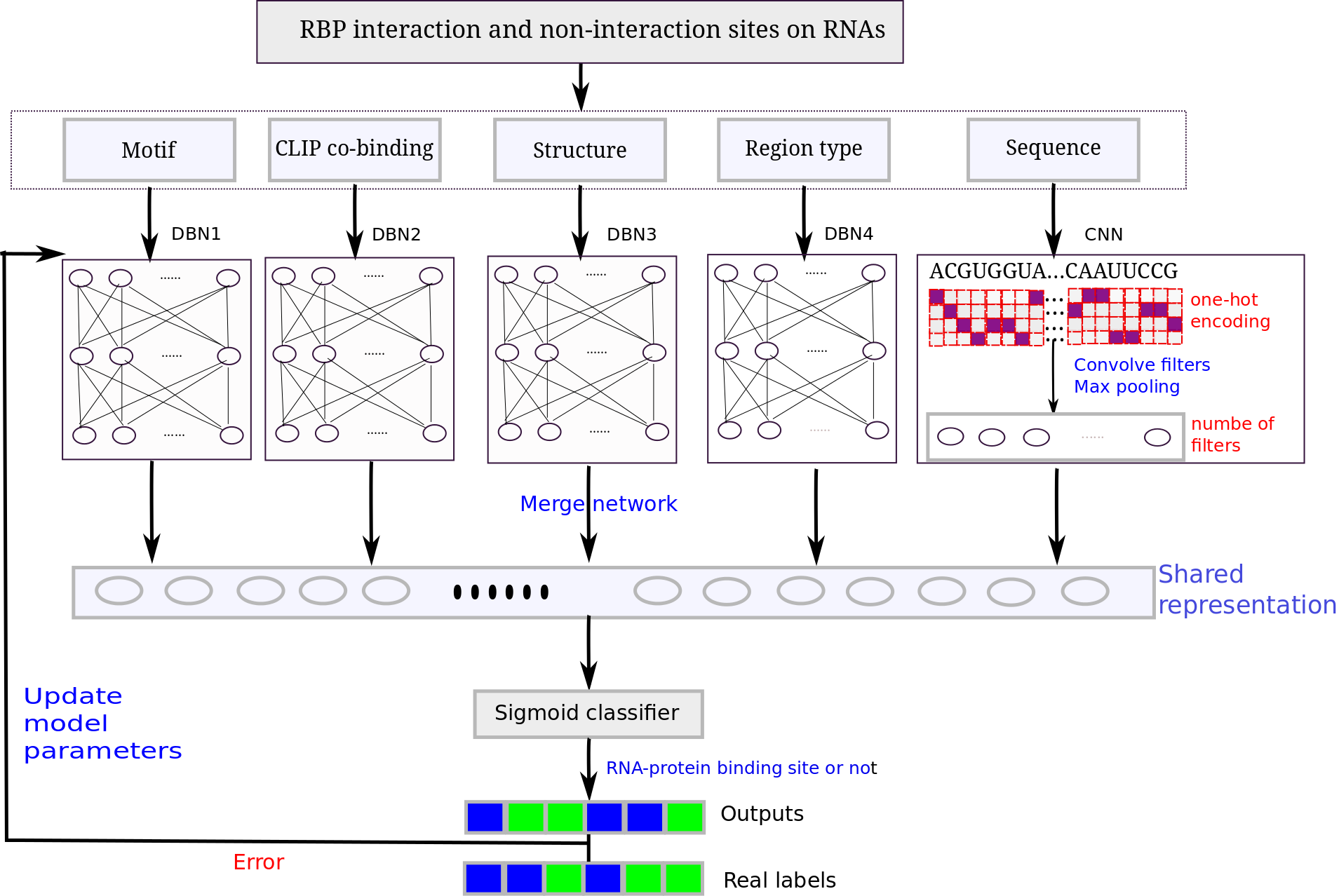
RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach
Prediction of binding property of RNA-binding proteins using multi-sized filters and multi-modal deep convolutional neural network | PLOS ONE

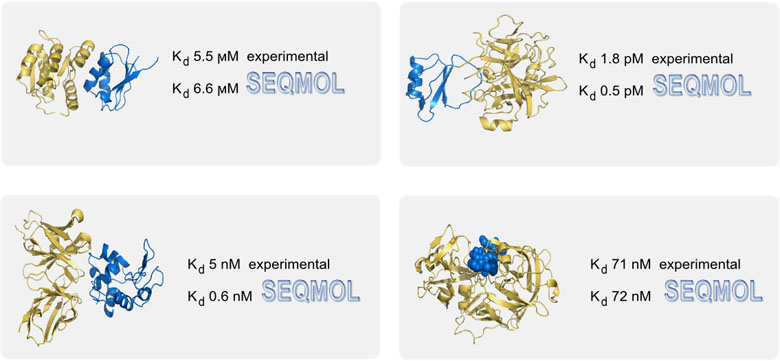
Interrogating RNA–Small Molecule Interactions with Structure Probing and Artificial Intelligence-Augmented Molecular Simulations | ACS Central Science

Computational approaches for the analysis of RNA–protein interactions: A primer for biologists - Journal of Biological Chemistry

The performance of RNA binding site prediction. (A) Comparison of ROC... | Download Scientific Diagram

RBPsuite: RNA-protein binding sites prediction suite based on deep learning | BMC Genomics | Full Text

HydRA: Deep-learning models for predicting RNA-binding capacity from protein interaction association context and protein sequence - ScienceDirect

RPI-Bind: a structure-based method for accurate identification of RNA-protein binding sites | Scientific Reports

Exploring the computational methods for protein-ligand binding site prediction - Computational and Structural Biotechnology Journal

DrugPred_RNA—A Tool for Structure-Based Druggability Predictions for RNA Binding Sites | Journal of Chemical Information and Modeling

Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks | BMC Genomics | Full Text
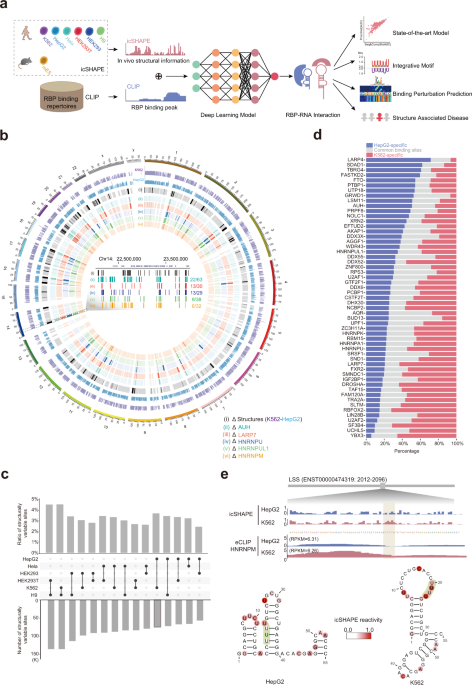
Predicting dynamic cellular protein–RNA interactions by deep learning using in vivo RNA structures | Cell Research

A deep learning framework to predict binding preference of RNA constituents on protein surface | Nature Communications

The workf low of RNA-small molecule binding site prediction. (A, B)... | Download Scientific Diagram

Two example of protein-RNA complexes (PDB id: 1I6U and 3IAB). (A) and... | Download Scientific Diagram