iDRBP_MMC: Identifying DNA-Binding Proteins and RNA-Binding Proteins Based on Multi-Label Learning Model and Motif-Based Convolutional Neural Network - ScienceDirect
![PDF] RNA-bioinformatics: Tools, services and databases for the analysis of RNA-based regulation. | Semantic Scholar PDF] RNA-bioinformatics: Tools, services and databases for the analysis of RNA-based regulation. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/9dece30b27fa1eafa5f54978806cd3a01ef0fef9/4-Figure1-1.png)
PDF] RNA-bioinformatics: Tools, services and databases for the analysis of RNA-based regulation. | Semantic Scholar
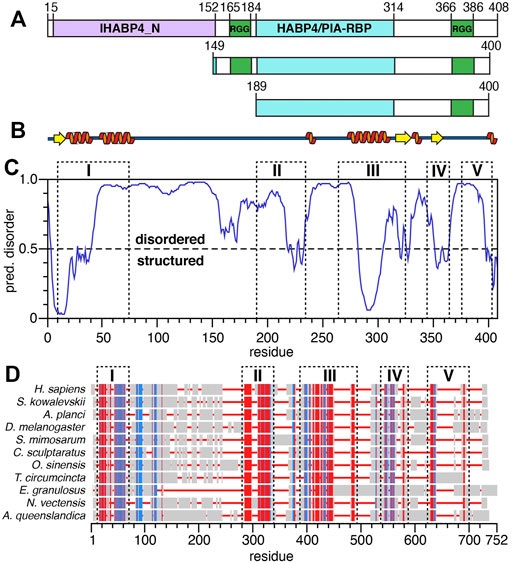
Frontiers | Structural Characterization of the RNA-Binding Protein SERBP1 Reveals Intrinsic Disorder and Atypical RNA Binding Modes

A deep learning framework to predict binding preference of RNA constituents on protein surface | Nature Communications
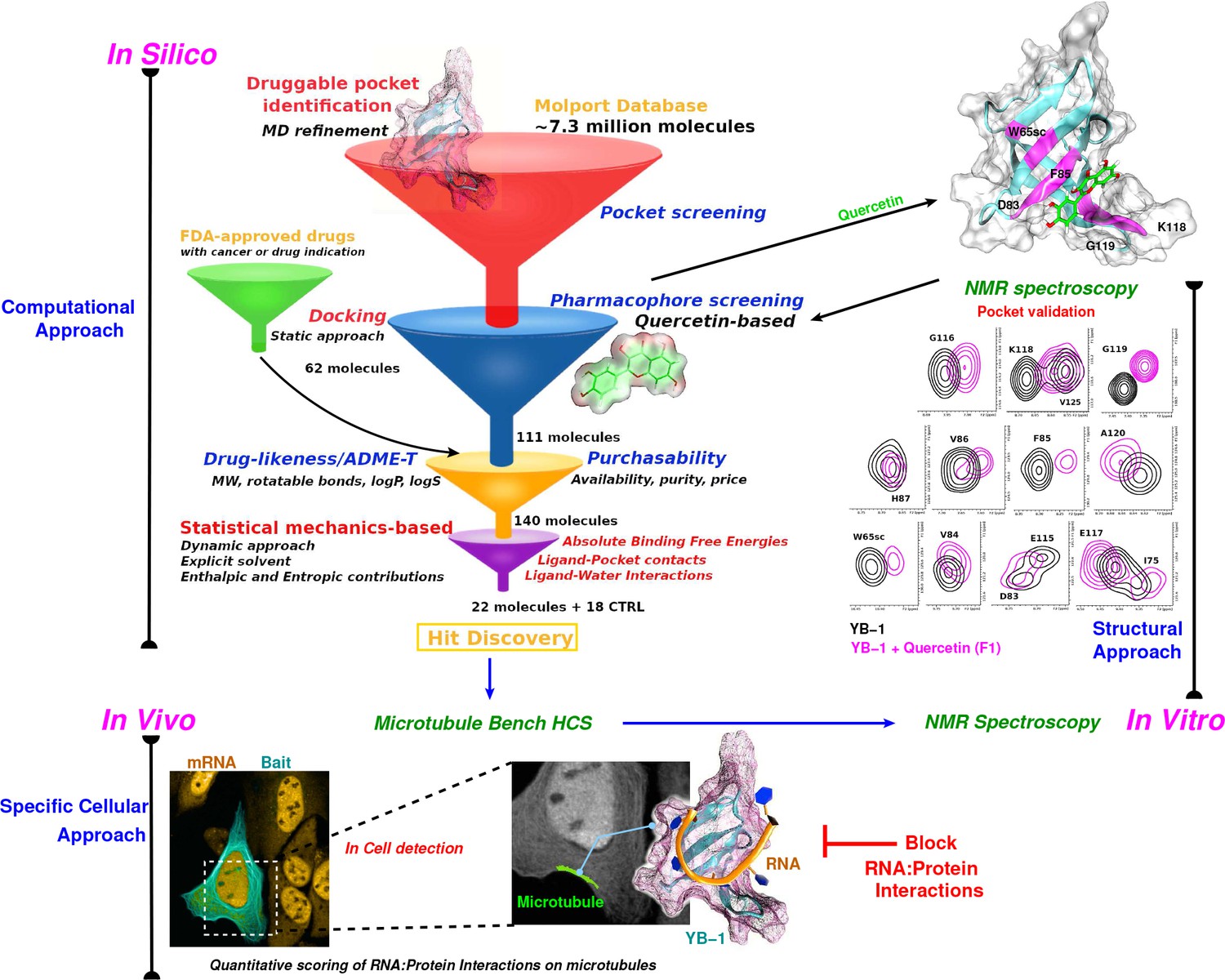
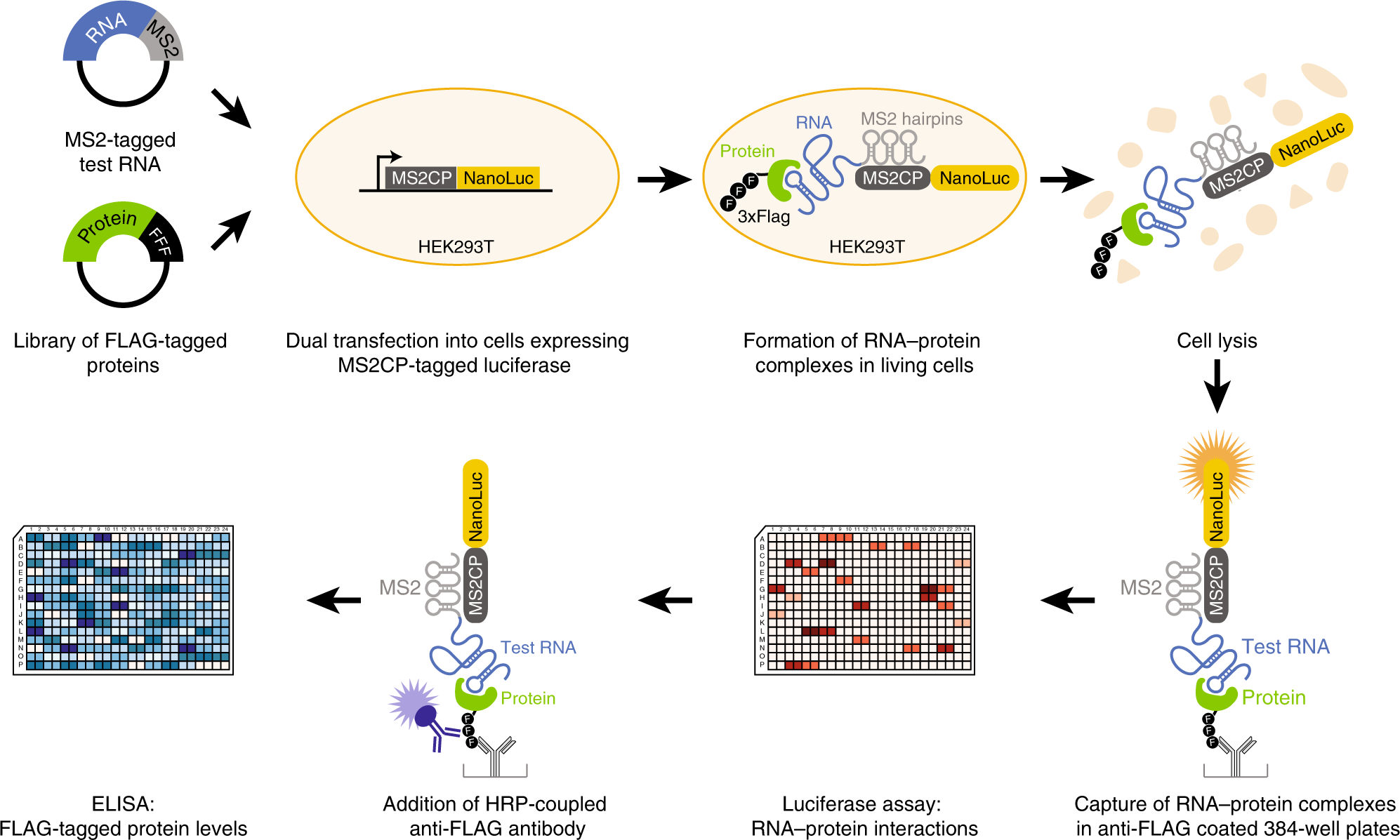
Targeting RNA:protein interactions with an integrative approach leads to the identification of potent YBX1 inhibitors | eLife

Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks | BMC Genomics | Full Text

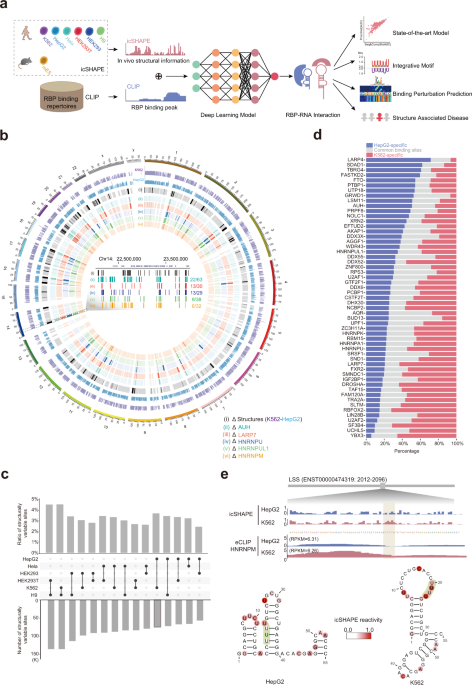
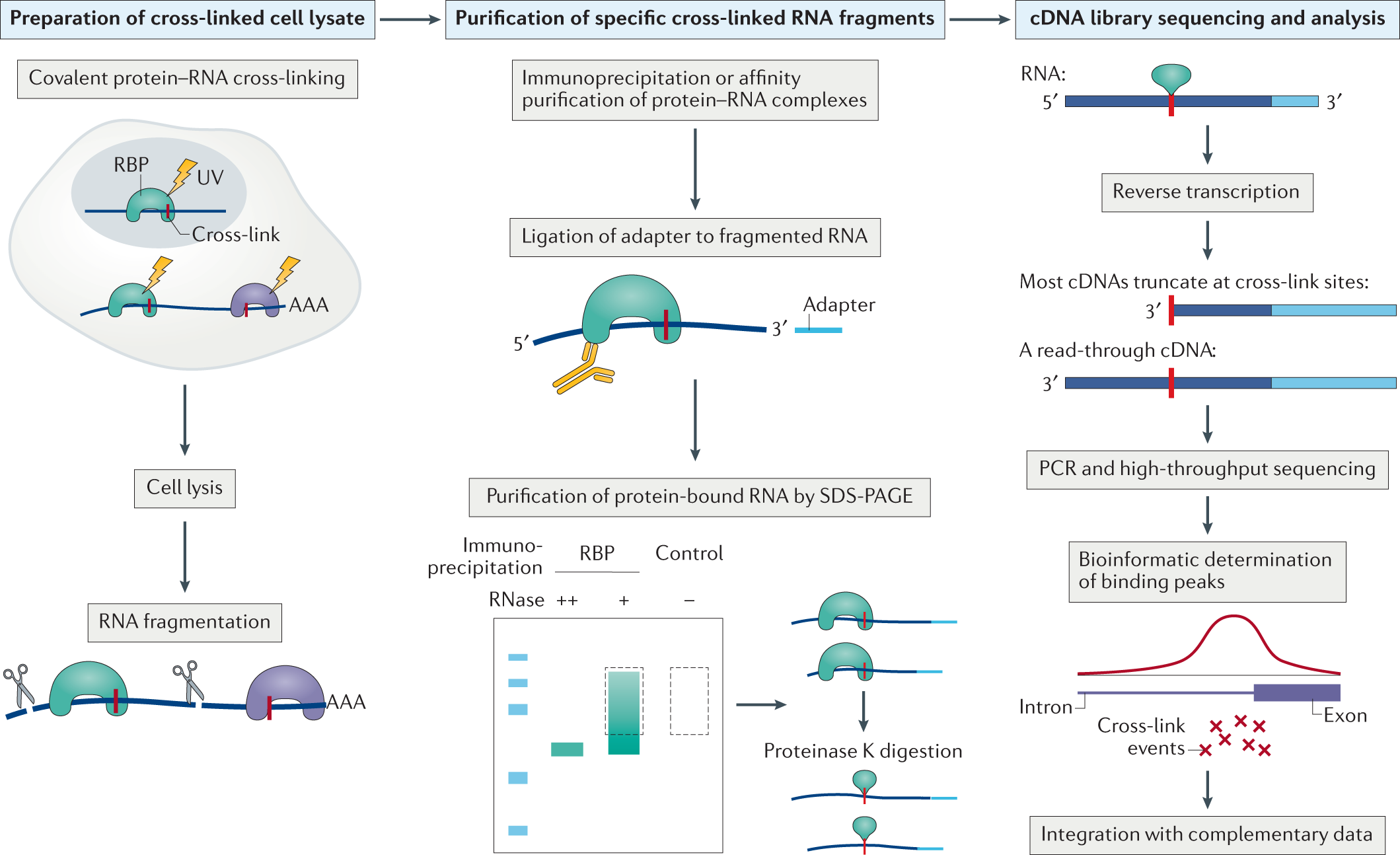
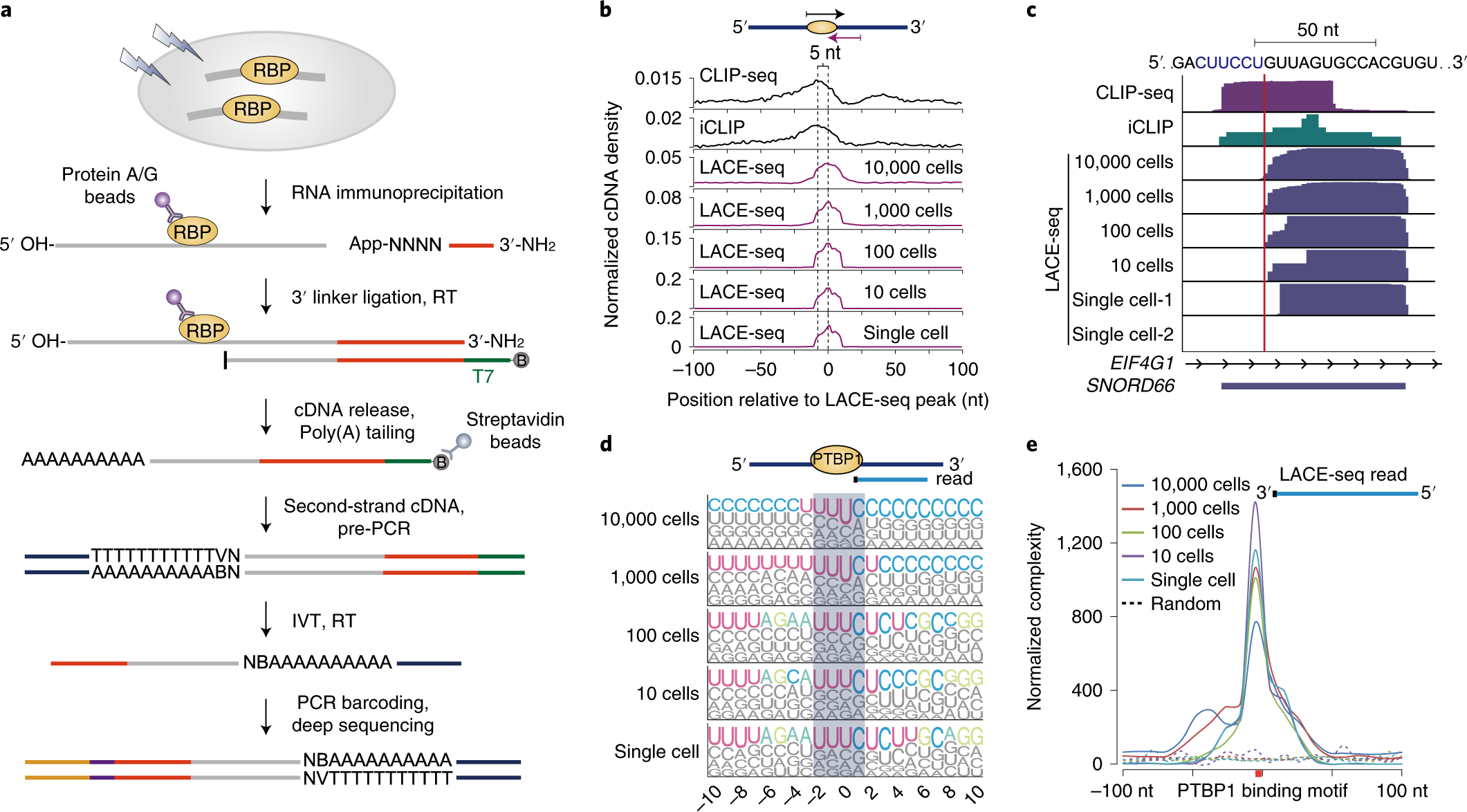
Predicting dynamic cellular protein–RNA interactions by deep learning using in vivo RNA structures | Cell Research

DrugPred_RNA—A Tool for Structure-Based Druggability Predictions for RNA Binding Sites | Journal of Chemical Information and Modeling
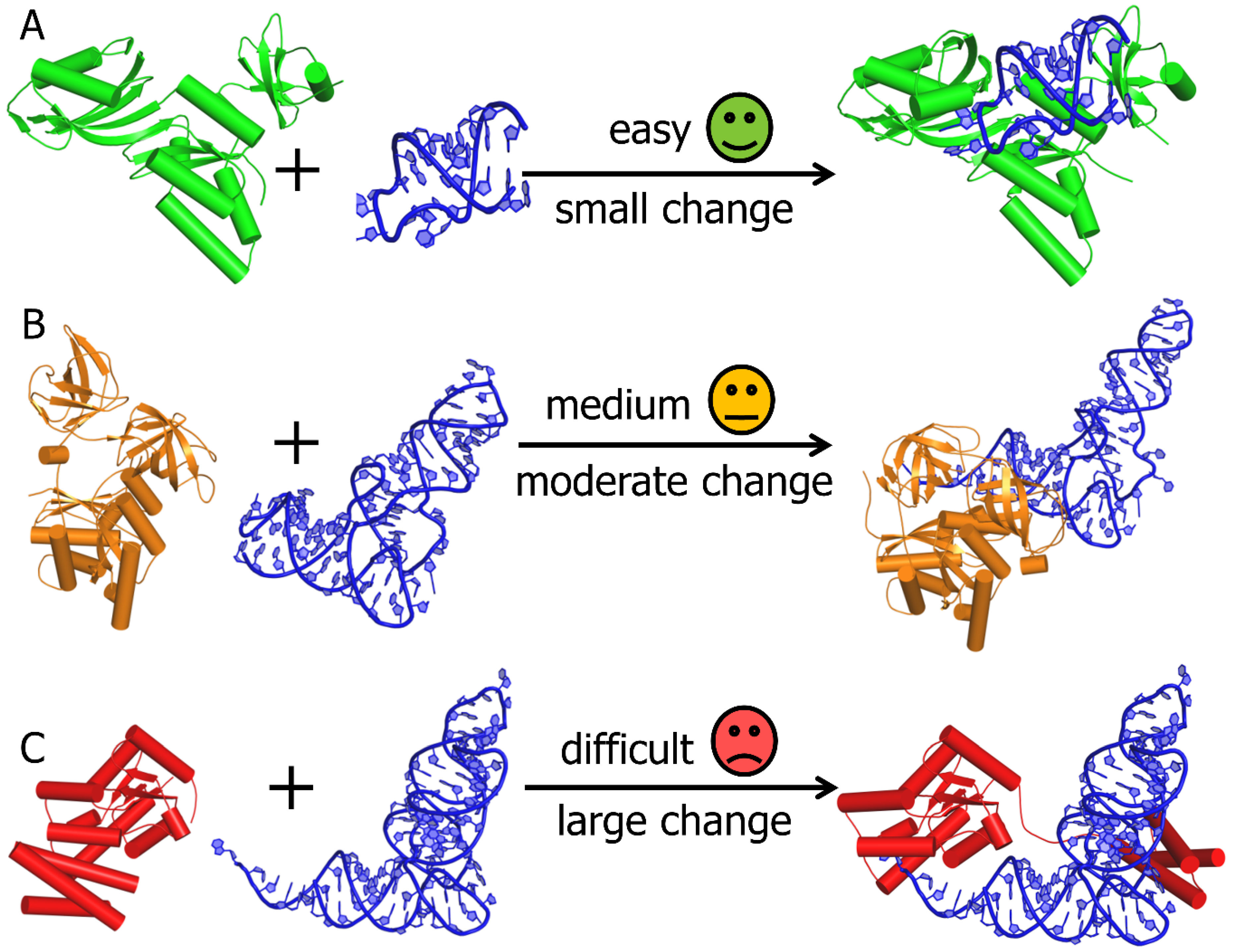
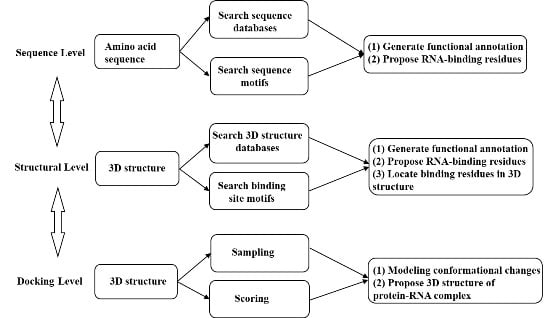
Genes | Free Full-Text | Bioinformatics Tools and Benchmarks for Computational Docking and 3D Structure Prediction of RNA-Protein Complexes

RBPsuite: RNA-protein binding sites prediction suite based on deep learning | BMC Genomics | Full Text
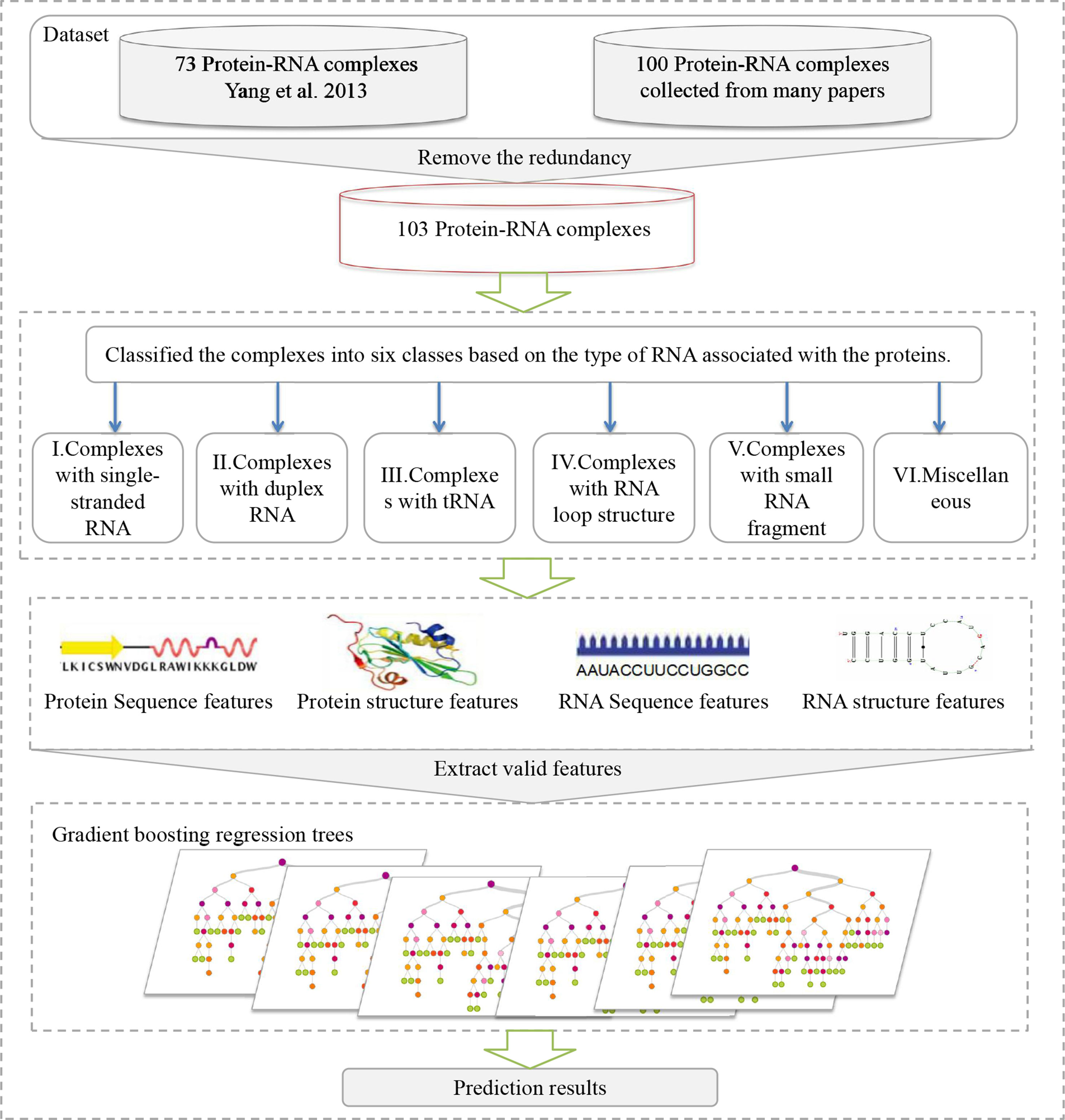
Frontiers | PredPRBA: Prediction of Protein-RNA Binding Affinity Using Gradient Boosted Regression Trees

Computational approaches for the analysis of RNA–protein interactions: A primer for biologists | Semantic Scholar

RNA-bioinformatics: Tools, services and databases for the analysis of RNA-based regulation - ScienceDirect

HydRA: Deep-learning models for predicting RNA-binding capacity from protein interaction association context and protein sequence - ScienceDirect