
The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis | Microbial Cell Factories | Full Text

21 The Ribosome Binding Site | Expression Engineering | Lecture 14 | Metabolic Engineering | SP20 - YouTube

The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis | Microbial Cell Factories | Full Text

Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA | PNAS

Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli | Scientific Reports

The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis | Microbial Cell Factories | Full Text
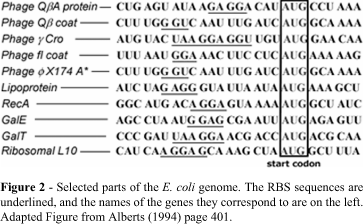
SciELO - Brasil - Ribosome binding site recognition using neural networks Ribosome binding site recognition using neural networks
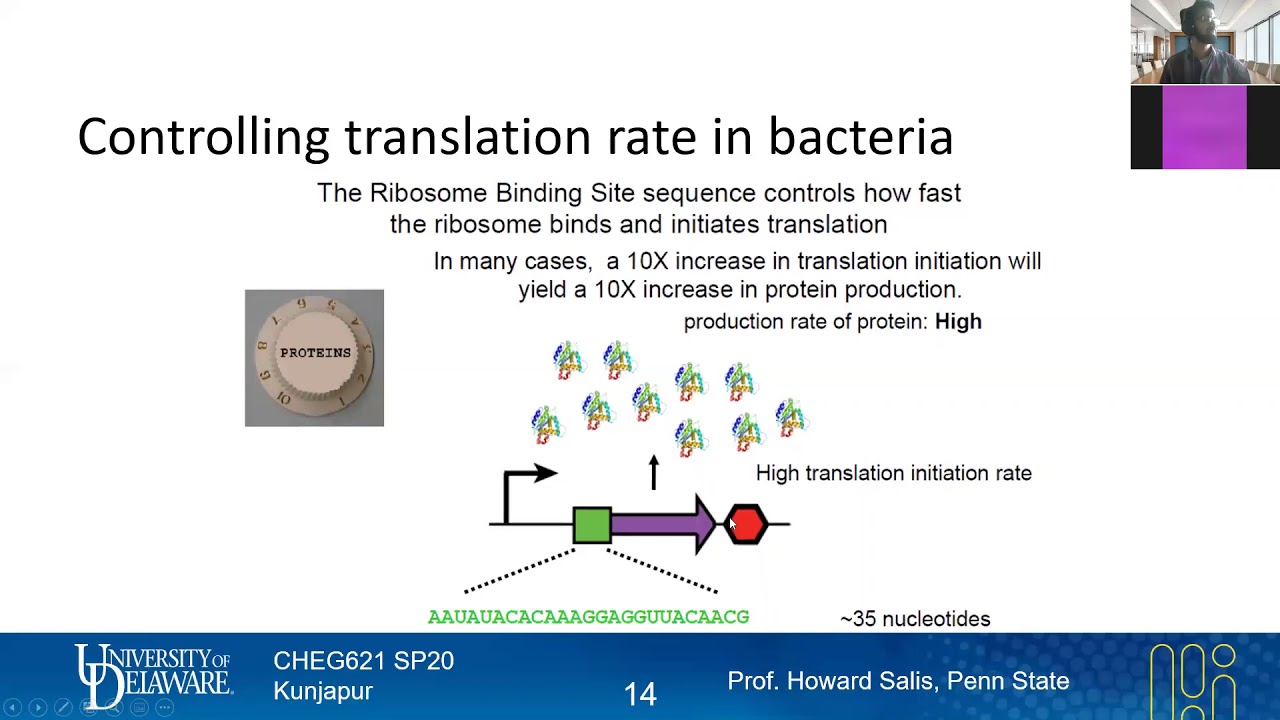
21 The Ribosome Binding Site | Expression Engineering | Lecture 14 | Metabolic Engineering | SP20 - YouTube
GitHub - hsalis/Ribosome-Binding-Site-Calculator-v1.0: The Ribosome Binding Site (RBS) Calculator can predict the translation initiation rate of a protein coding sequence in bacteria and design synthetic RBS sequences to rationally control the translation
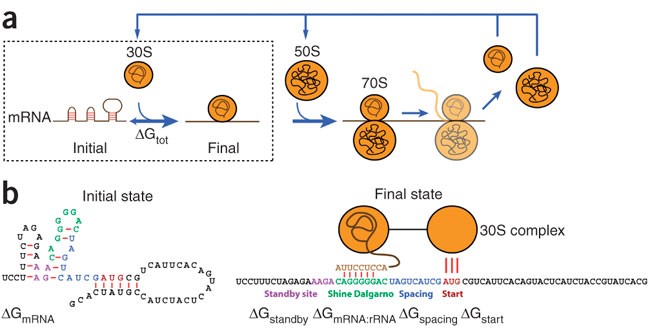
Automated design of synthetic ribosome binding sites to control protein expression | Nature Biotechnology

The context of the ribosome binding site in mRNAs defines specificity of action of kasugamycin, an inhibitor of translation initiation | PNAS
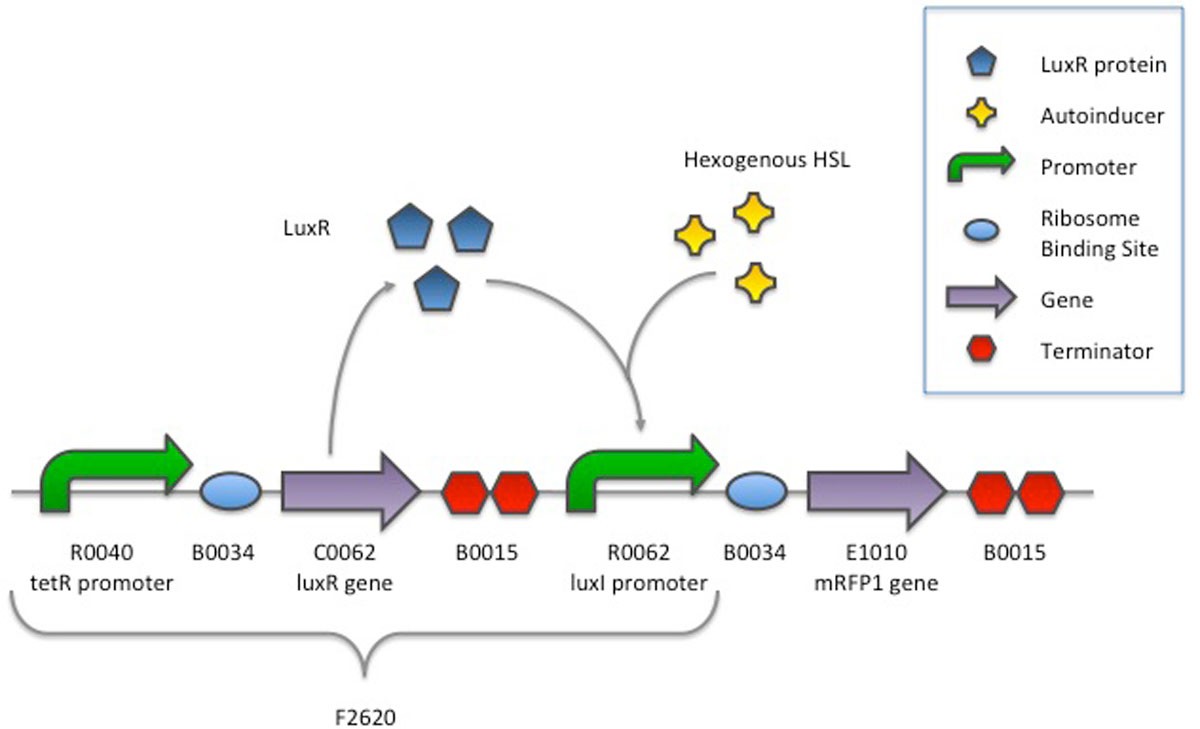
Characterization of an inducible promoter in different DNA copy number conditions | BMC Bioinformatics | Full Text
How many tRNA binding sites are there on ribosomes? My textbook (NCERT) says 2 and the internet says 3. - Quora

Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli | Scientific Reports

DrugPred_RNA—A Tool for Structure-Based Druggability Predictions for RNA Binding Sites | Journal of Chemical Information and Modeling
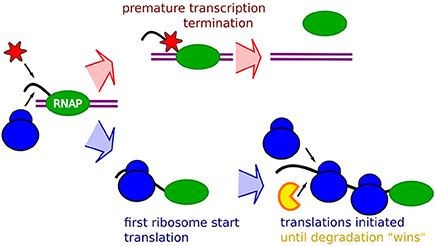
Frontiers | Occlusion of the Ribosome Binding Site Connects the Translational Initiation Frequency, mRNA Stability and Premature Transcription Termination

Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli | Scientific Reports
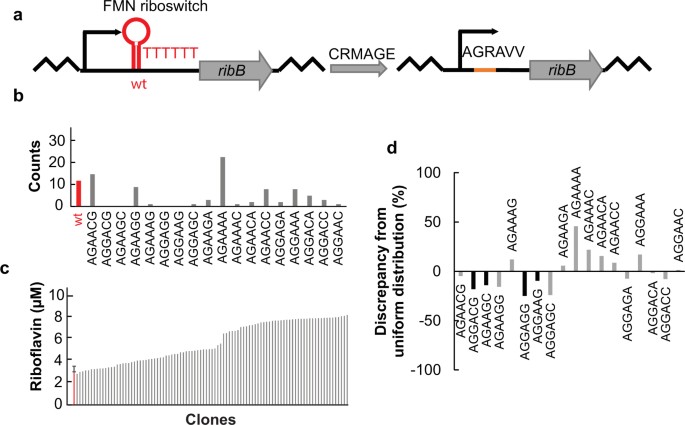
Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli | Scientific Reports

Efficient search, mapping, and optimization of multi‐protein genetic systems in diverse bacteria | Molecular Systems Biology



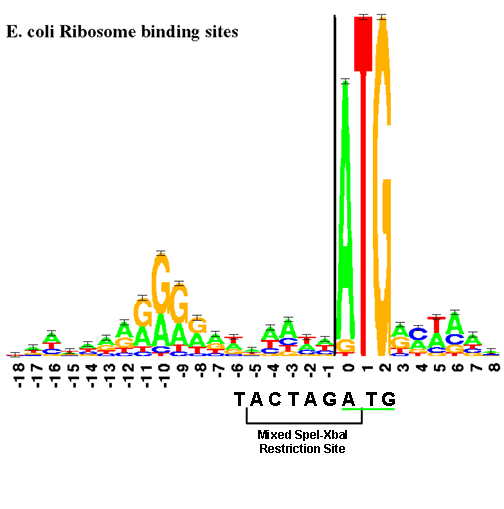

![PDF] Classification of Escherichia coli K-12 ribosome binding sites | Semantic Scholar PDF] Classification of Escherichia coli K-12 ribosome binding sites | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/4af616123bce83b1f5dff449961c1df697c8d771/2-Figure2-1.png)
