A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection - ScienceDirect
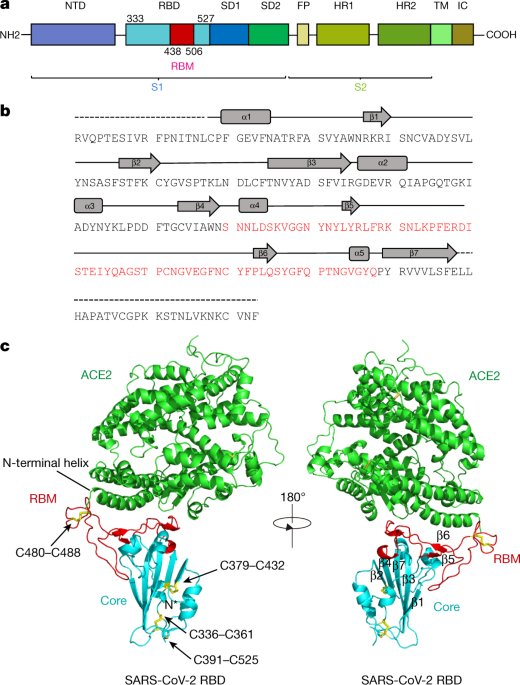
Viruses | Free Full-Text | Structural Analysis of Neutralizing Epitopes of the SARS-CoV-2 Spike to Guide Therapy and Vaccine Design Strategies
![2019 Coronavirus SARS-CoV-2 Receptor Binding Motif [LT5578] - $499.00 : LifeTein, The Peptide Synthesis Service Company 2019 Coronavirus SARS-CoV-2 Receptor Binding Motif [LT5578] - $499.00 : LifeTein, The Peptide Synthesis Service Company](https://www.lifetein.com/images/peptide-product-images/Coronavirus-RBM.png)
2019 Coronavirus SARS-CoV-2 Receptor Binding Motif [LT5578] - $499.00 : LifeTein, The Peptide Synthesis Service Company
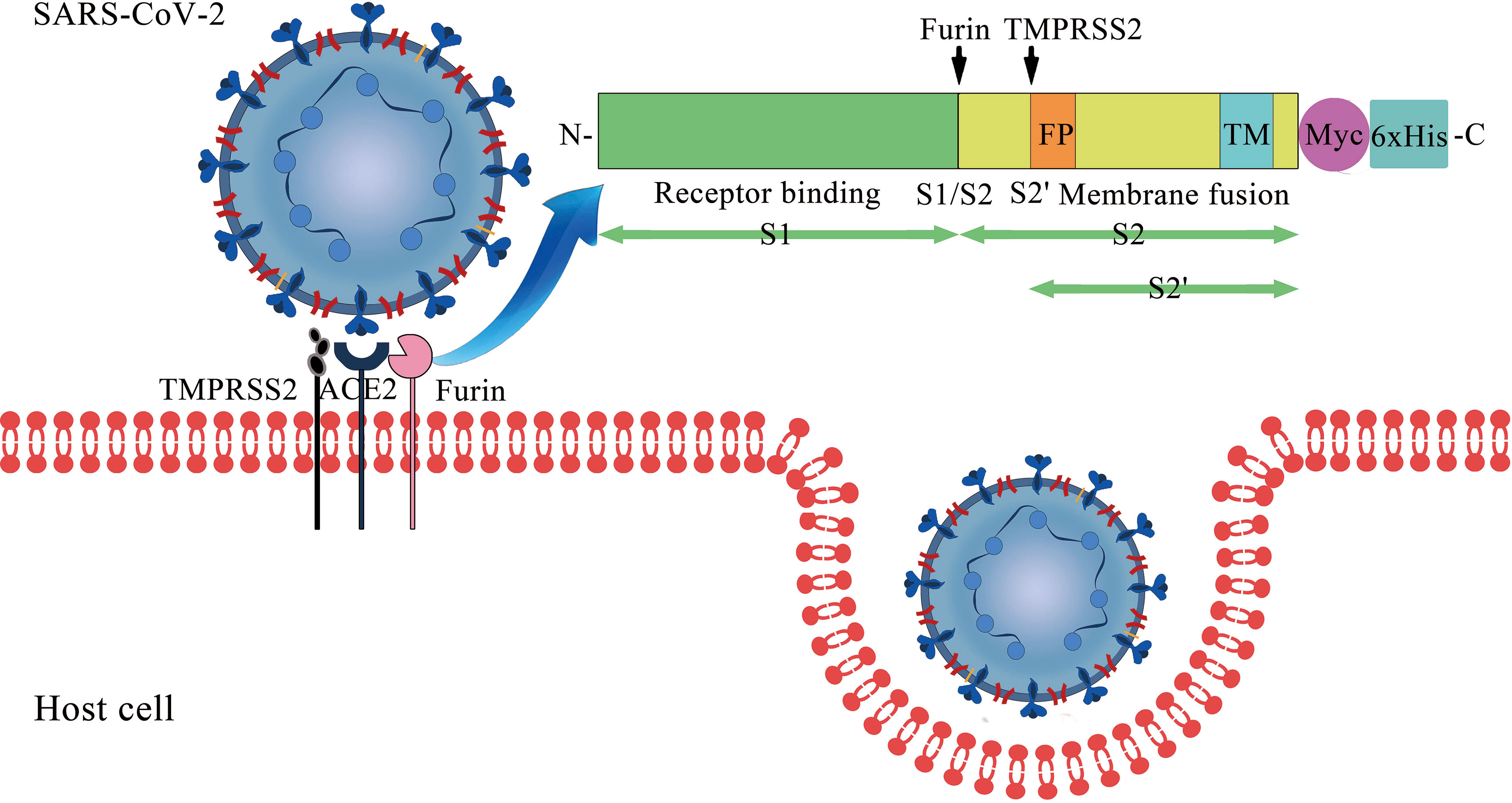
Frontiers | The Impact of ACE2 Polymorphisms on COVID-19 Disease: Susceptibility, Severity, and Therapy
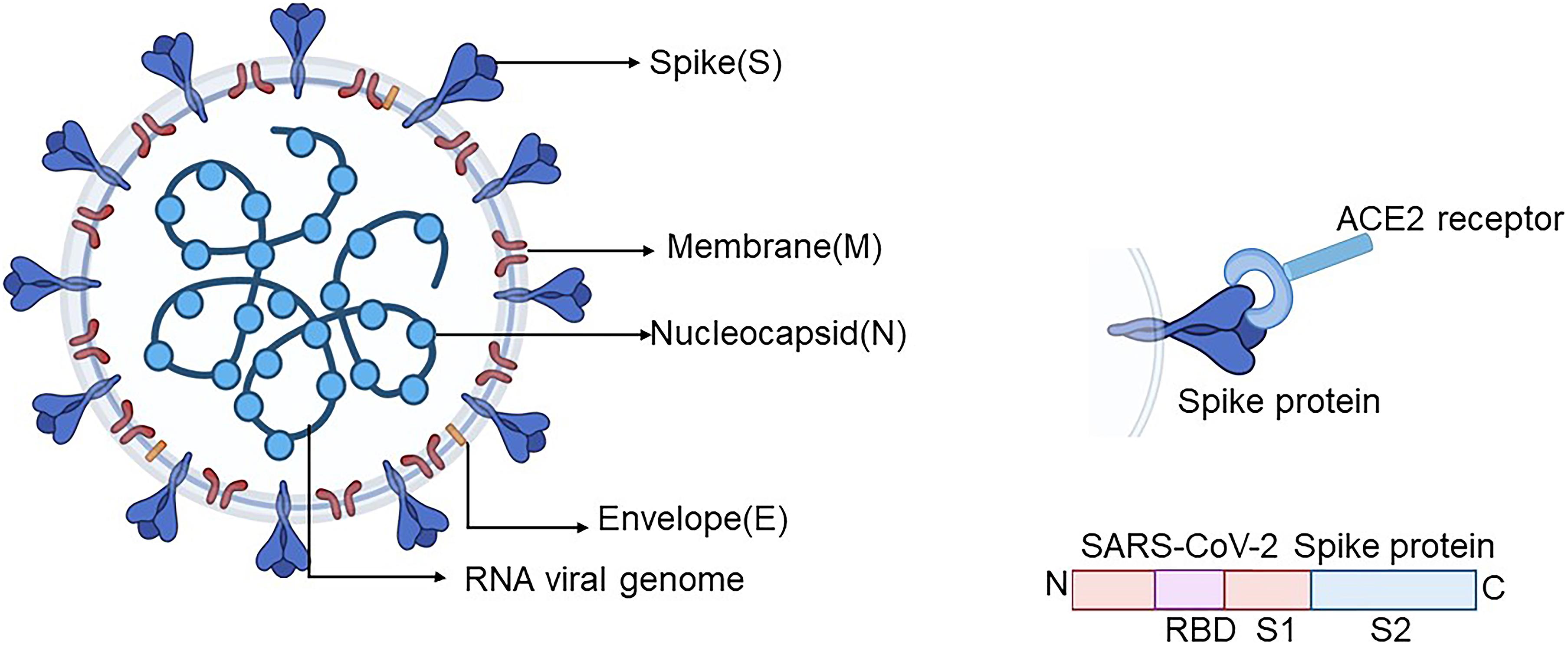
3-D Structure of SARS-CoV-2 Explains High Infectivity vs. Other Coronaviruses | Advanced Photon Source
Molecular recognition of SARS-CoV-2 spike glycoprotein: quantum chemical hot spot and epitope analyses - Chemical Science (RSC Publishing) DOI:10.1039/D0SC06528E
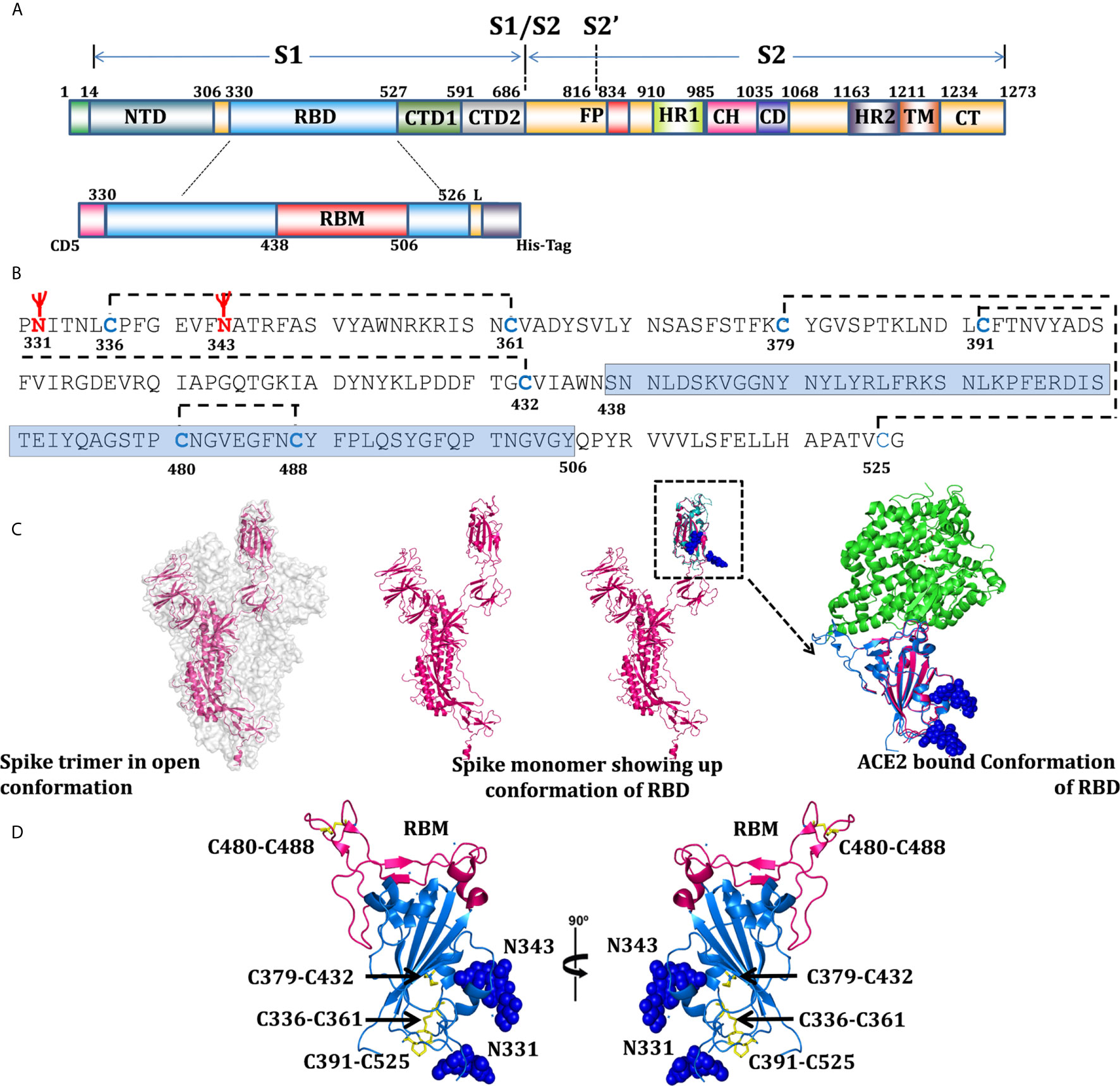
Frontiers | Comparative Immunomodulatory Evaluation of the Receptor Binding Domain of the SARS-CoV-2 Spike Protein; a Potential Vaccine Candidate Which Imparts Potent Humoral and Th1 Type Immune Response in a Mouse Model

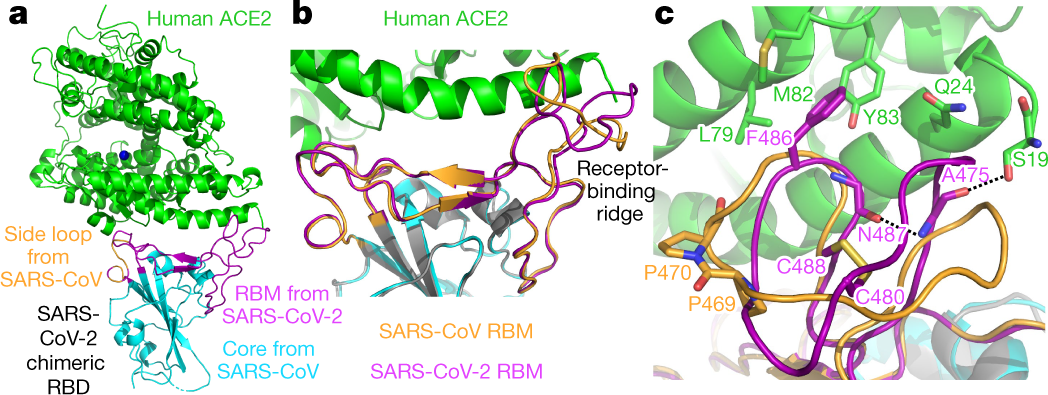
Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor | Nature Communications

CIMB | Free Full-Text | The Occluded Epitope Residing in Spike Receptor-Binding Motif Is Essential for Cross-Neutralization of SARS-CoV-2 Delta Variant

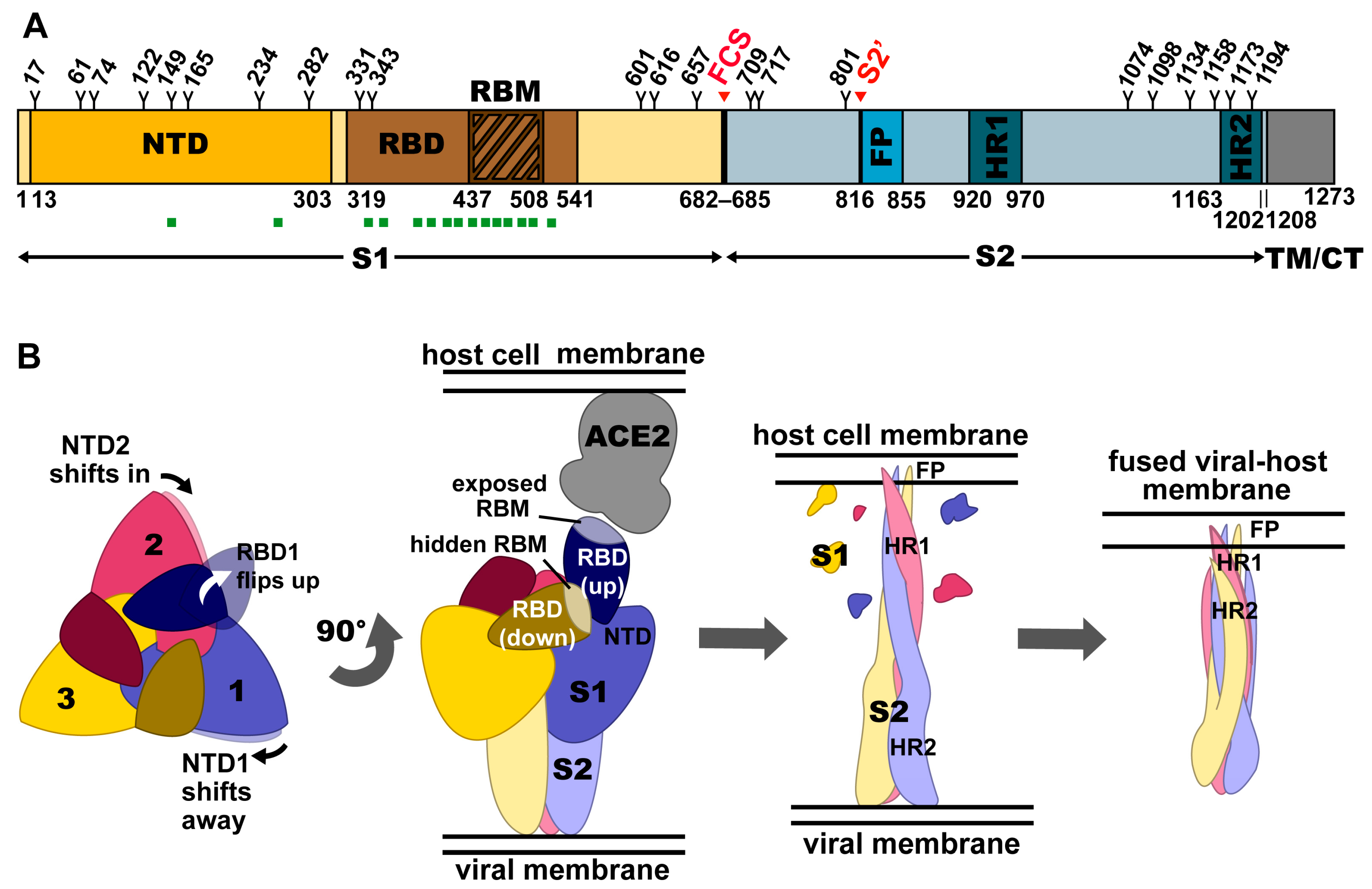
A comparative study of human betacoronavirus spike proteins: structure, function and therapeutics | Archives of Virology

Chromosome-Wide Mapping of Estrogen Receptor Binding Reveals Long-Range Regulation Requiring the Forkhead Protein FoxA1: Cell

Predicted structural mimicry of spike receptor-binding motifs from highly pathogenic human coronaviruses - ScienceDirect

Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV | bioRxiv

Is the Rigidity of SARS-CoV-2 Spike Receptor-Binding Motif the Hallmark for Its Enhanced Infectivity? Insights from All-Atom Simulations | The Journal of Physical Chemistry Letters

Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies | Cellular & Molecular Immunology

Targeting the Receptor-Binding Motif of SARS-CoV-2 with D-Peptides Mimicking the ACE2 Binding Helix: Lessons for Inhibiting Omicron and Future Variants of Concern | Journal of Chemical Information and Modeling

Protein structure of the receptor binding motif (RBM) of the receptor... | Download Scientific Diagram

Mutations in the receptor-binding motif (RBM) of the omicron variant.... | Download Scientific Diagram

Predicted structural mimicry of spike receptor-binding motifs from highly pathogenic human coronaviruses - Computational and Structural Biotechnology Journal