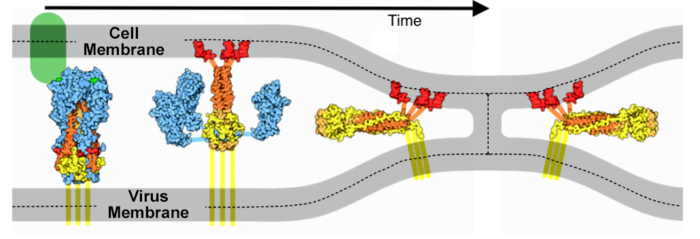
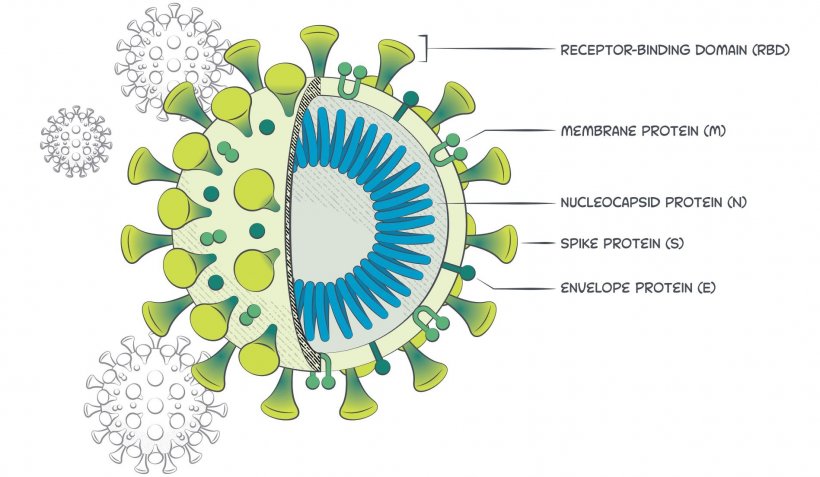
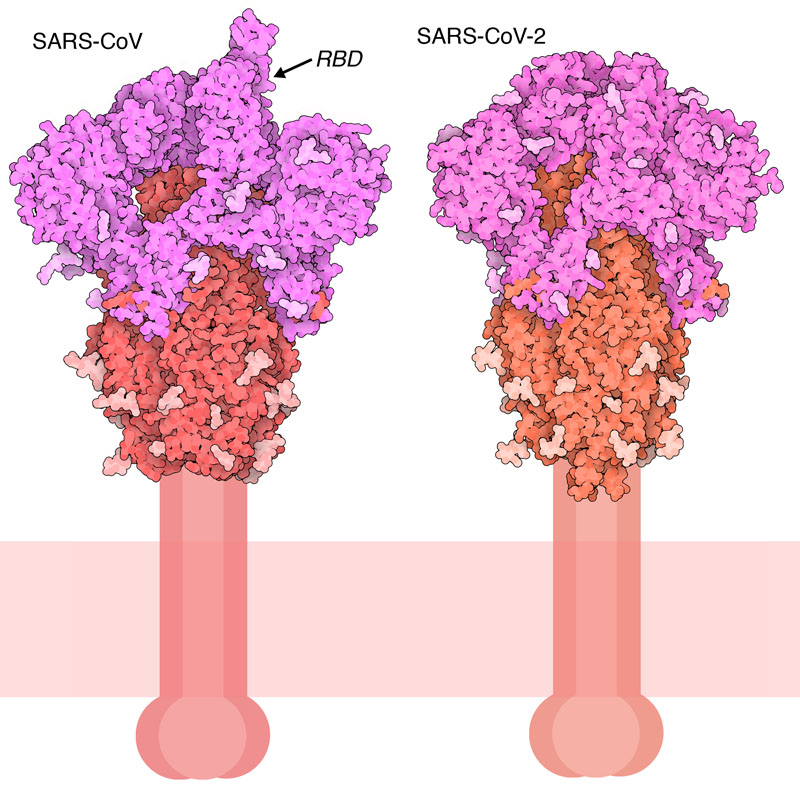
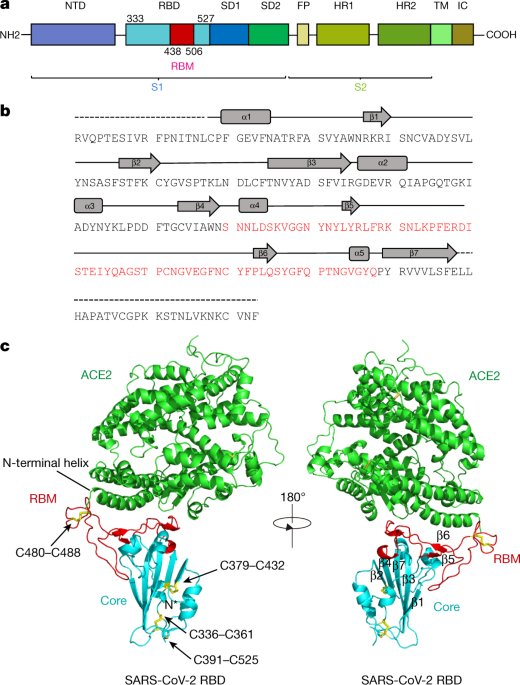
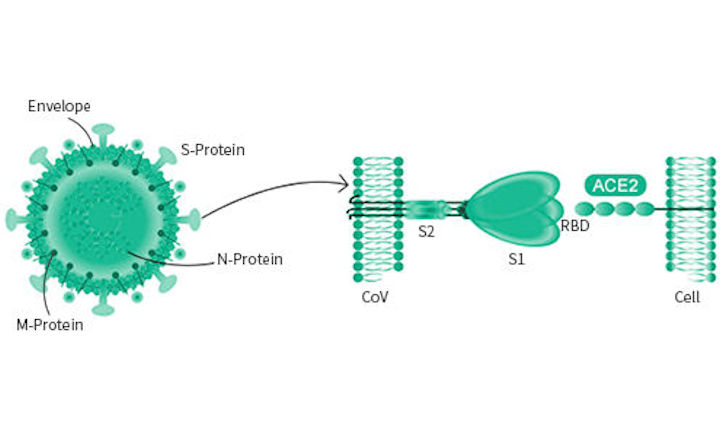
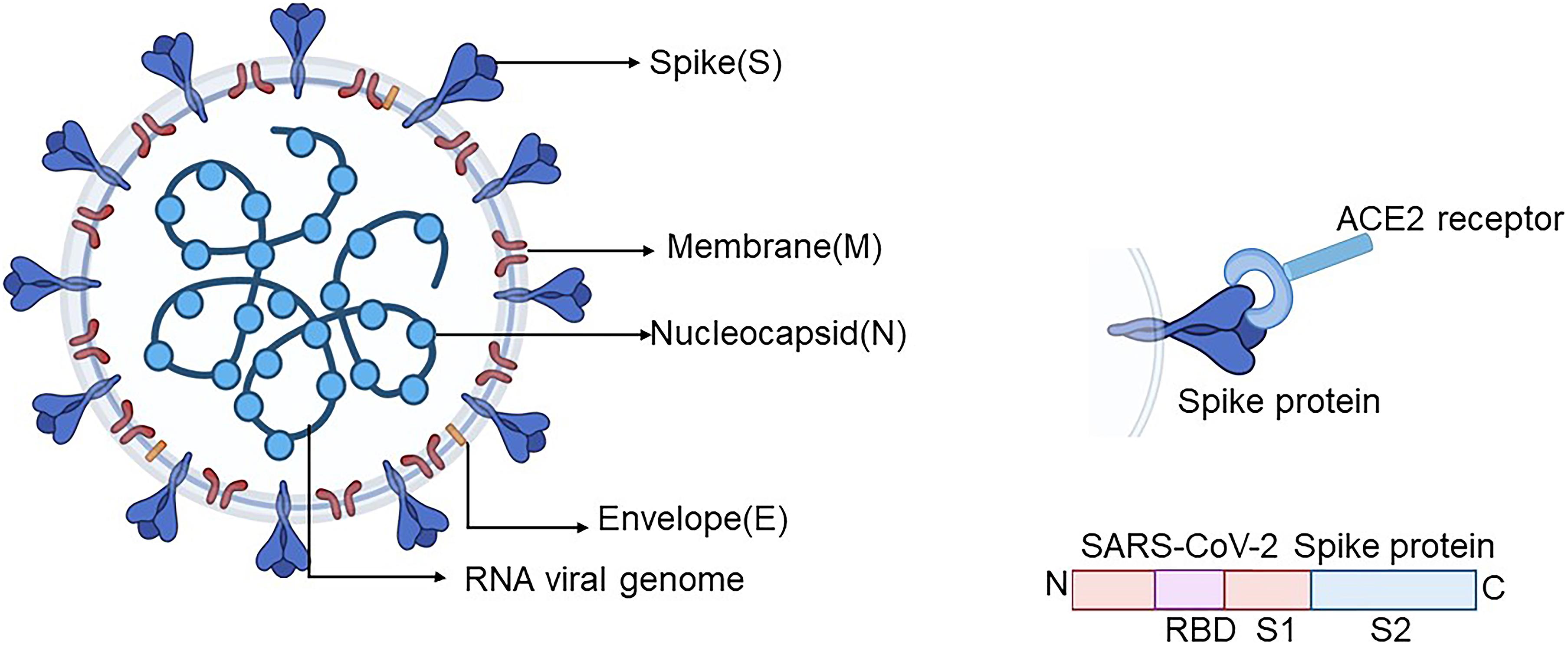
In Silico Structure-Based Repositioning of Approved Drugs for Spike Glycoprotein S2 Domain Fusion Peptide of SARS-CoV-2: Rationale from Molecular Dynamics and Binding Free Energy Calculations | mSystems

Differences and similarities between SARS-CoV and SARS-CoV-2: spike receptor -binding domain recognition and host cell infection with support of cellular serine proteases | Infection

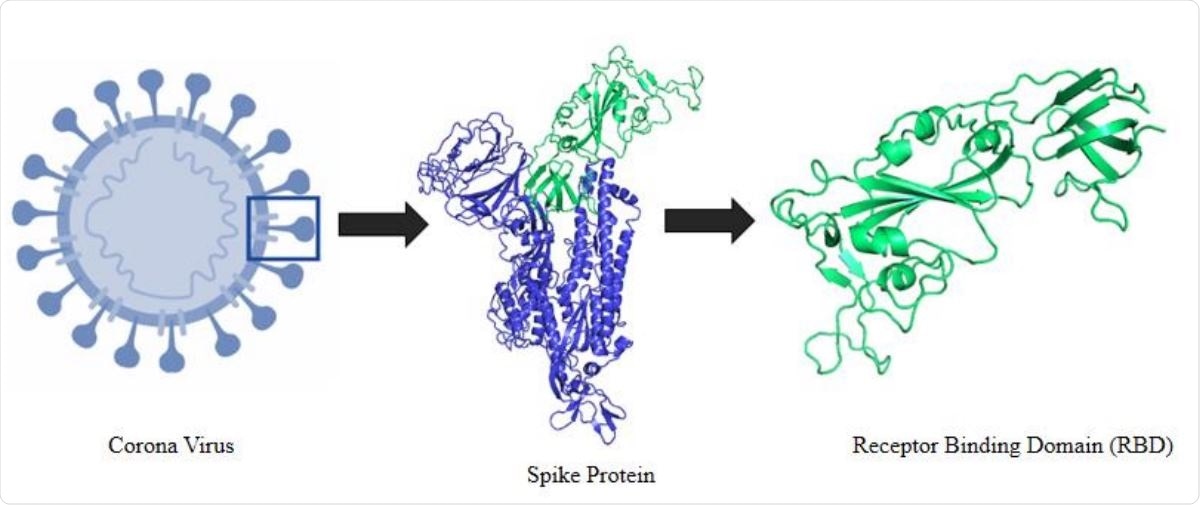
a) Structure of the receptor-binding domain (RBD) of the S protein in... | Download Scientific Diagram

SARS-CoV-2 IgG antibodies and why the receptor-binding domain of the spike protein is so important - YouTube

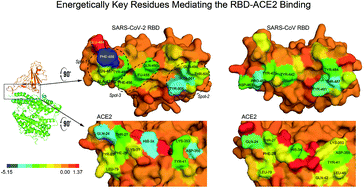
Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity - Computational and Structural Biotechnology Journal
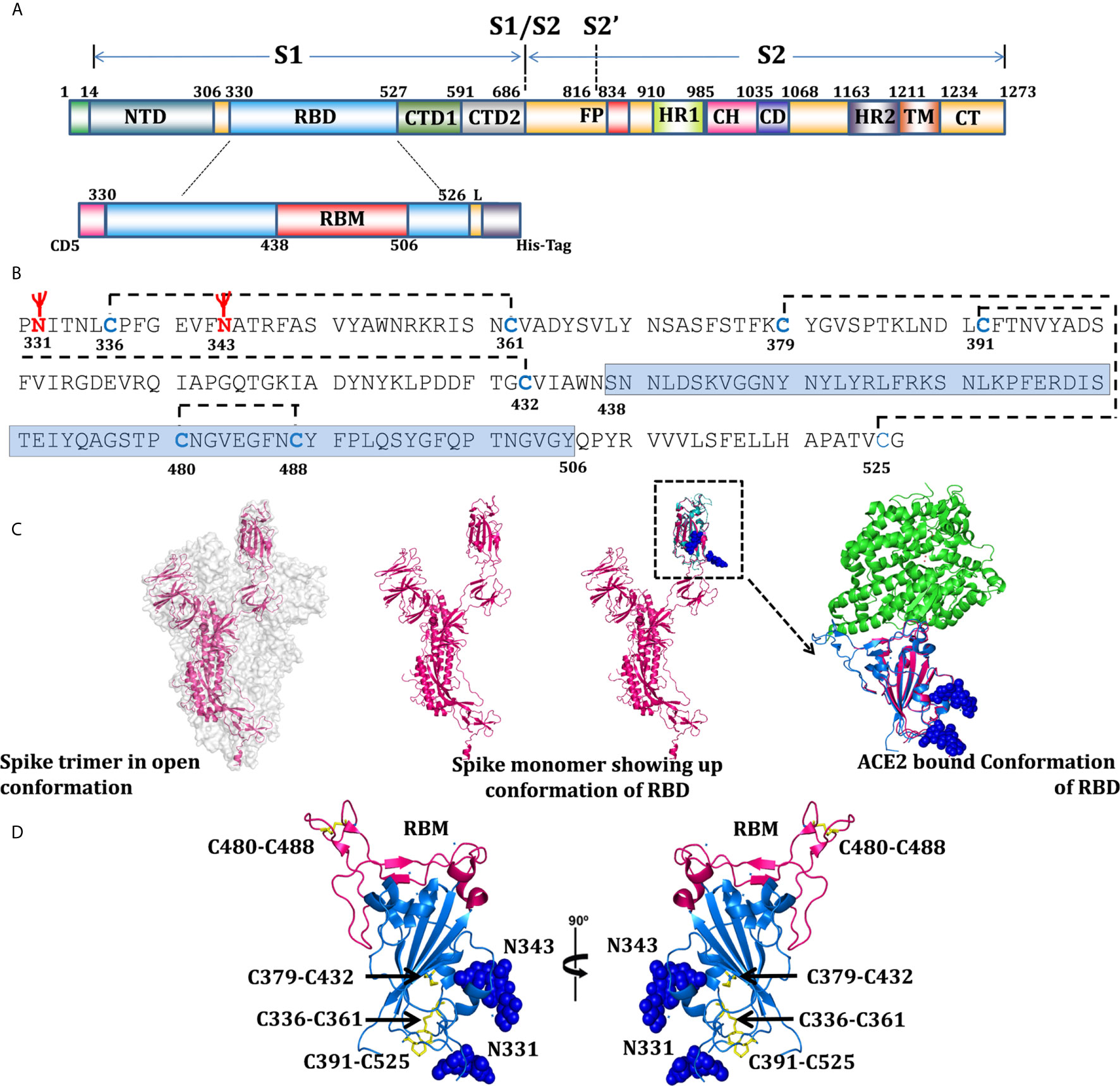
Frontiers | Comparative Immunomodulatory Evaluation of the Receptor Binding Domain of the SARS-CoV-2 Spike Protein; a Potential Vaccine Candidate Which Imparts Potent Humoral and Th1 Type Immune Response in a Mouse Model
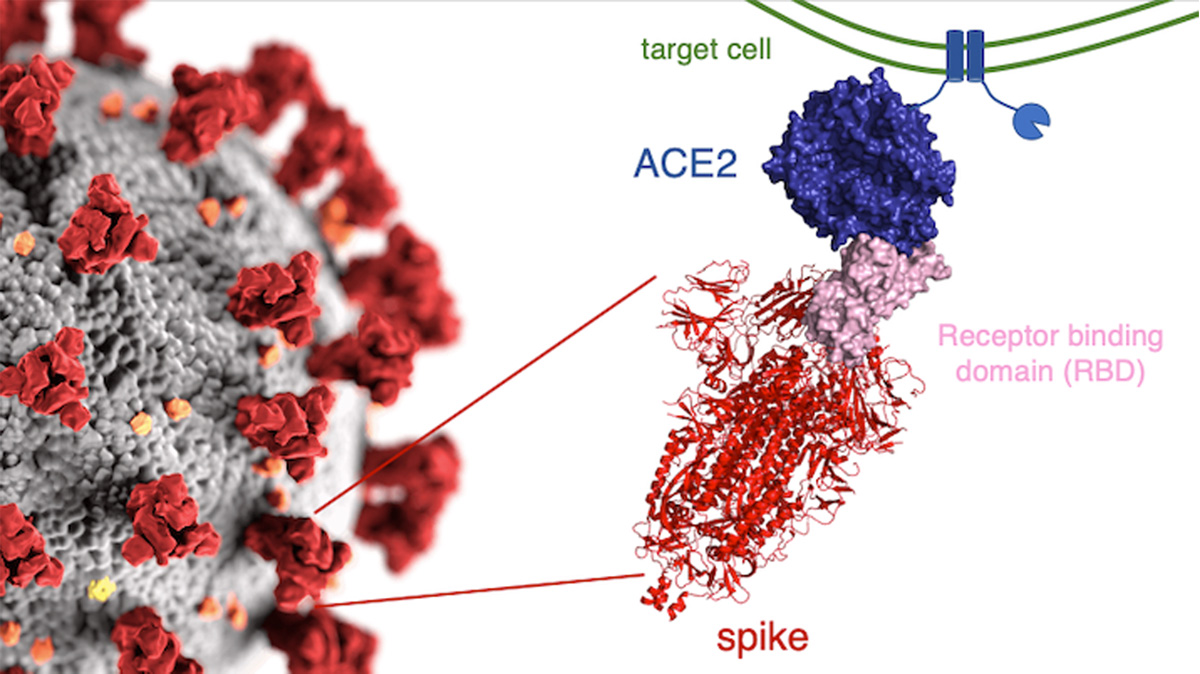
Map Catalogs Effects of Coronavirus Mutations- Crop Biotech Update (September 2, 2020) | Crop Biotech Update - ISAAA.org

Computational screening of 645 antiviral peptides against the receptor-binding domain of the spike protein in SARS-CoV-2 - ScienceDirect
Construction and immunogenic studies of a mFc fusion receptor binding domain (RBD) of spike protein as a subunit vaccine against SARS-CoV-2 infection - Chemical Communications (RSC Publishing)
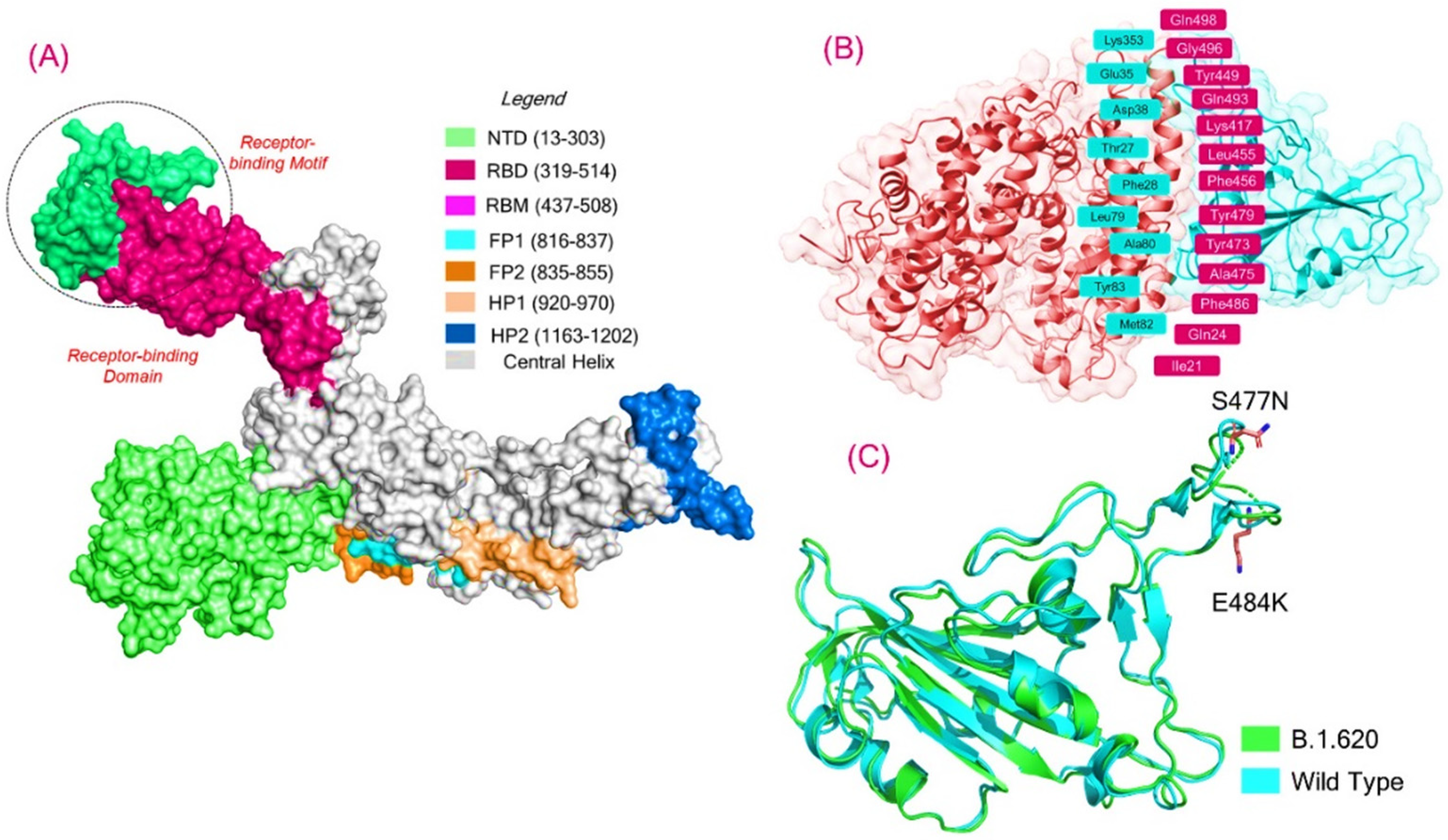
Biology | Free Full-Text | Insights into the Binding of Receptor-Binding Domain (RBD) of SARS-CoV-2 Wild Type and B.1.620 Variant with hACE2 Using Molecular Docking and Simulation Approaches

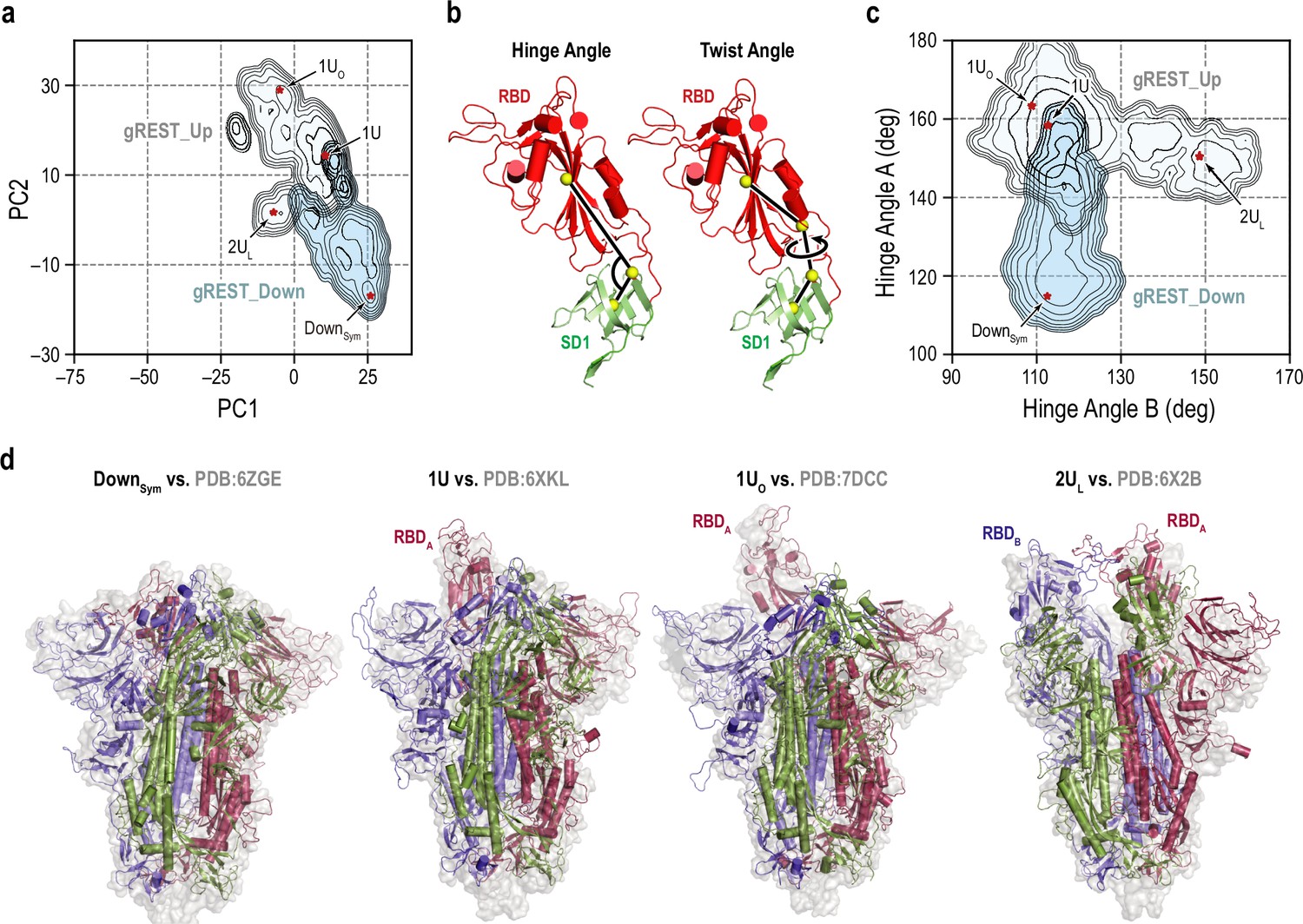
Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains - ScienceDirect