SARS-CoV-2 Omicron Variant: ACE2 Binding, Cryo-EM Structure of Spike Protein-ACE2 Complex and Antibody Evasion | bioRxiv
Frontiers | Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies

Characterization of SARS-CoV-2 Omicron spike RBD reveals significantly decreased stability, severe evasion of neutralizing-antibody recognition but unaffected engagement by decoy ACE2 modified for enhanced RBD binding | Signal Transduction and Targeted ...

Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor - ScienceDirect

SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2 | Signal Transduction and Targeted Therapy
Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains | PLOS Pathogens
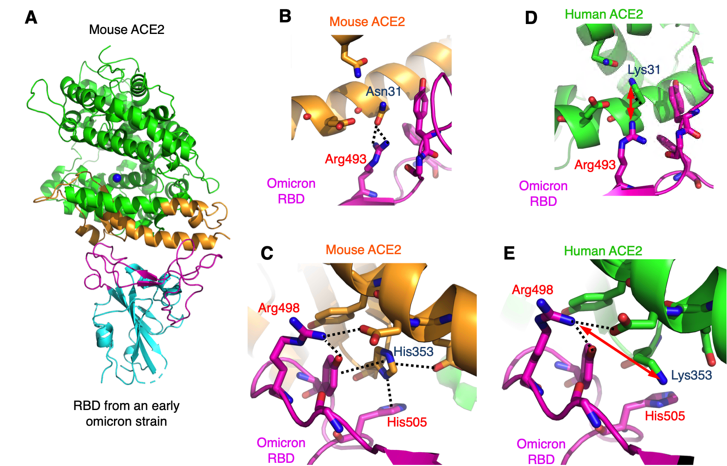
Structural Evidence that Rodents Facilitated the Evolution of the SARS-CoV-2 Omicron Variant | Stanford Synchrotron Radiation Lightsource
![Dynamics of the interaction between the receptor-binding domain of SARS-CoV-2 Omicron (B.1.1.529) variant and human angiotensin-converting enzyme 2 [PeerJ] Dynamics of the interaction between the receptor-binding domain of SARS-CoV-2 Omicron (B.1.1.529) variant and human angiotensin-converting enzyme 2 [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2022/13680/1/fig-1-full.png)
Dynamics of the interaction between the receptor-binding domain of SARS-CoV-2 Omicron (B.1.1.529) variant and human angiotensin-converting enzyme 2 [PeerJ]

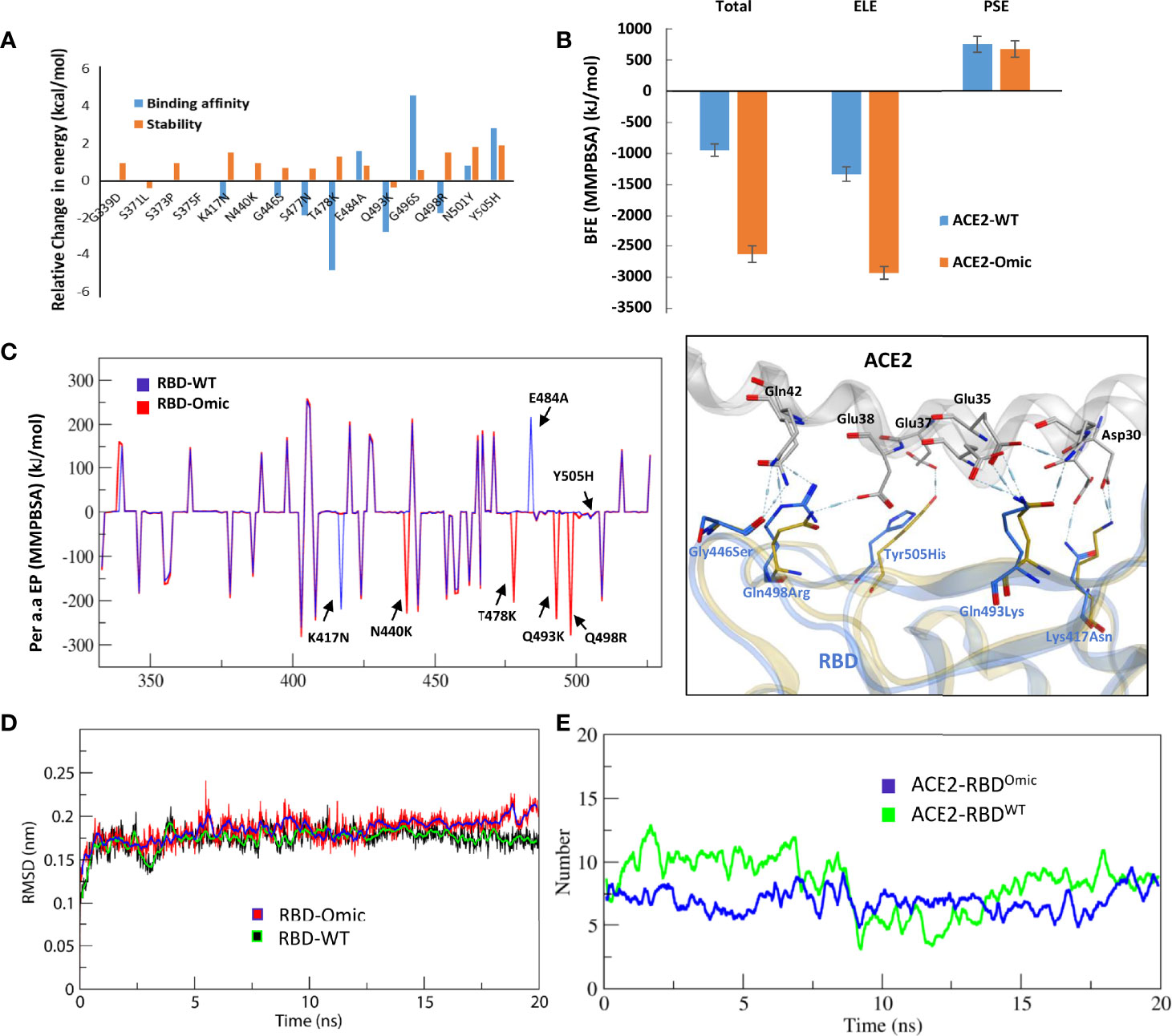
Probing conformational landscapes of binding and allostery in the SARS-CoV-2 omicron variant complexes using microsecond atomistic simulations and perturbation-based profiling approaches: hidden role of omicron mutations as modulators of allosteric ...

Targeting the Receptor-Binding Motif of SARS-CoV-2 with D-Peptides Mimicking the ACE2 Binding Helix: Lessons for Inhibiting Omicron and Future Variants of Concern | Journal of Chemical Information and Modeling

Biology | Free Full-Text | Targeting the RBD of Omicron Variant (B.1.1.529) with Medicinal Phytocompounds to Abrogate the Binding of Spike Glycoprotein with the hACE2 Using Computational Molecular Search and Simulation Approach

Binding affinity of ACE2 to the RBDs of WT, Delta and Omicron variants.... | Download Scientific Diagram

Characterization of the binding affinity between mouse ACE2 and Omicron... | Download Scientific Diagram
IJMS | Free Full-Text | The Increased Amyloidogenicity of Spike RBD and pH-Dependent Binding to ACE2 May Contribute to the Transmissibility and Pathogenic Properties of SARS-CoV-2 Omicron as Suggested by In Silico

Antibody binding and ACE2 binding inhibition is significantly reduced for the Omicron variant compared to all other variants of concern | medRxiv

Exploring the Binding Affinity and Mechanism between ACE2 and the Trimers of Delta and Omicron Spike Proteins by Molecular Dynamics Simulation and Bioassay | Journal of Chemical Information and Modeling
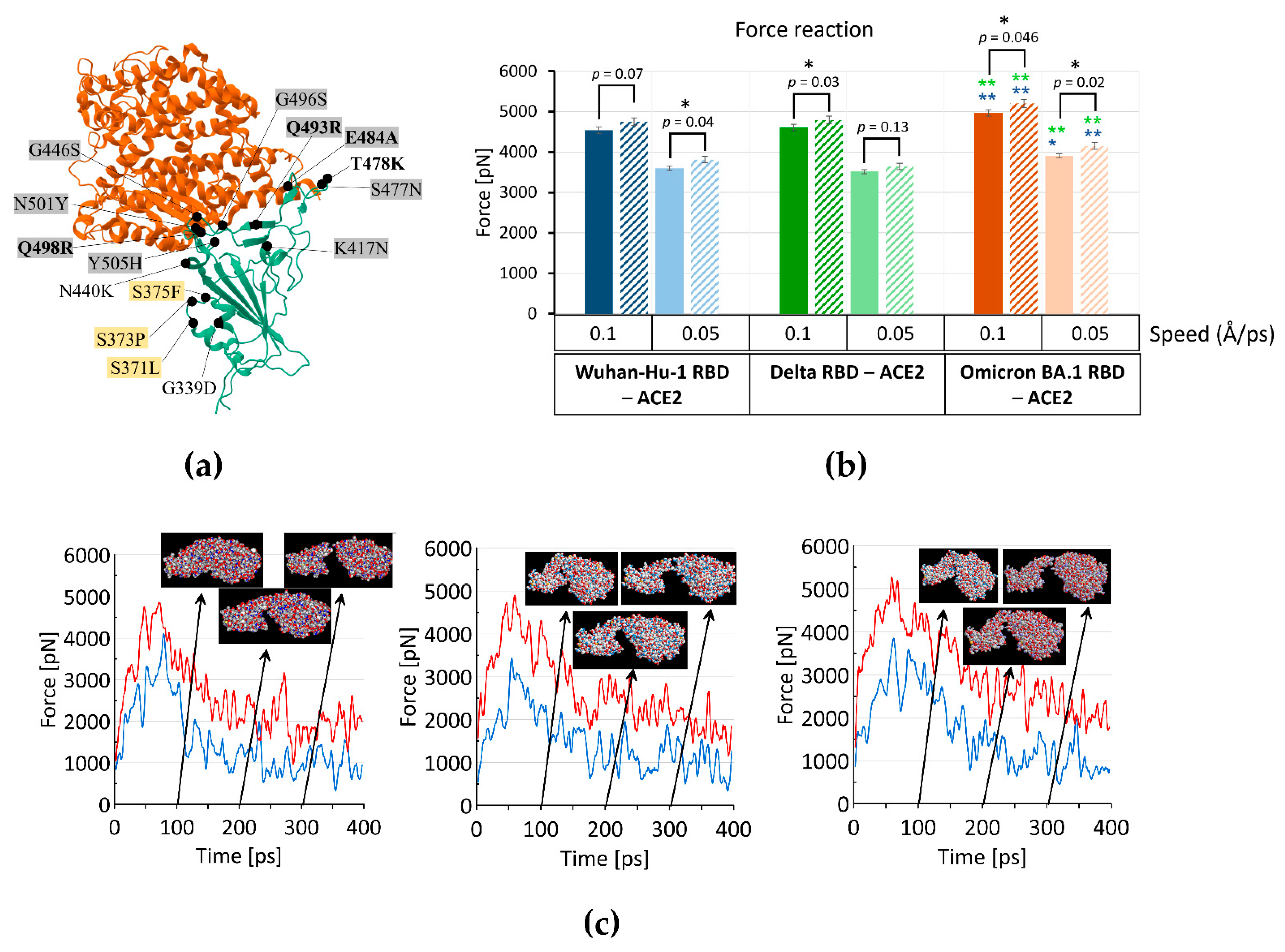
Frontiers | Deciphering the Impact of Mutations on the Binding Efficacy of SARS-CoV-2 Omicron and Delta Variants With Human ACE2 Receptor

Significance of the RBD mutations in the SARS-CoV-2 Omicron: from spike opening to antibody escape and cell attachment | bioRxiv

Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant | BMC Medicine | Full Text

The effect of the multiple mutations in Omicron RBD on its binding to human ACE2 receptor and immune evasion: an investigation of molecular dynamics simulations | Theoretical and Computational Chemistry | ChemRxiv

Omicron: Master of immune evasion maintains robust ACE2 binding | Signal Transduction and Targeted Therapy