PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci | BMC Bioinformatics | Full Text

Pioneer Transcription Factors Target Partial DNA Motifs on Nucleosomes to Initiate Reprogramming - ScienceDirect

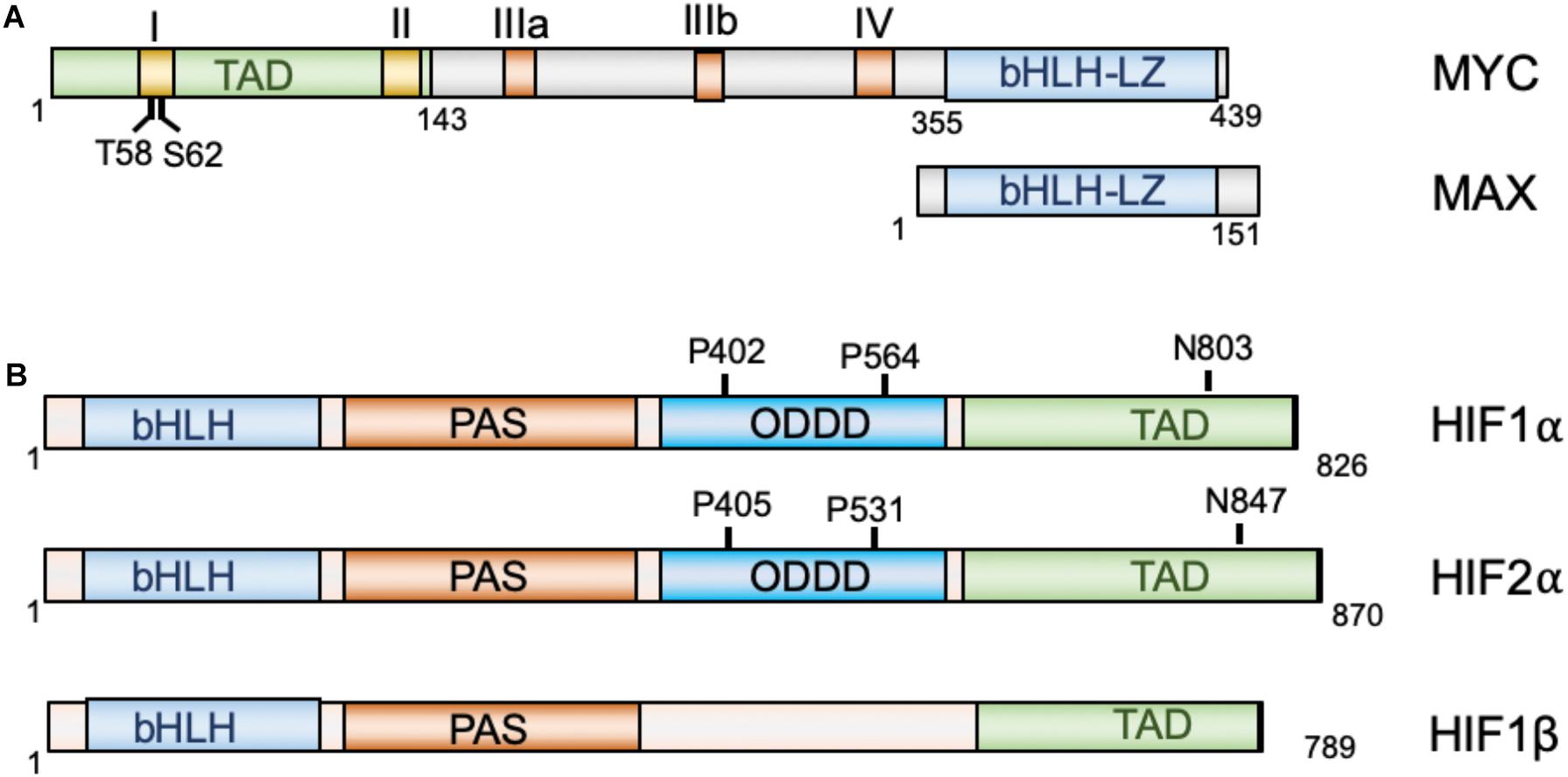
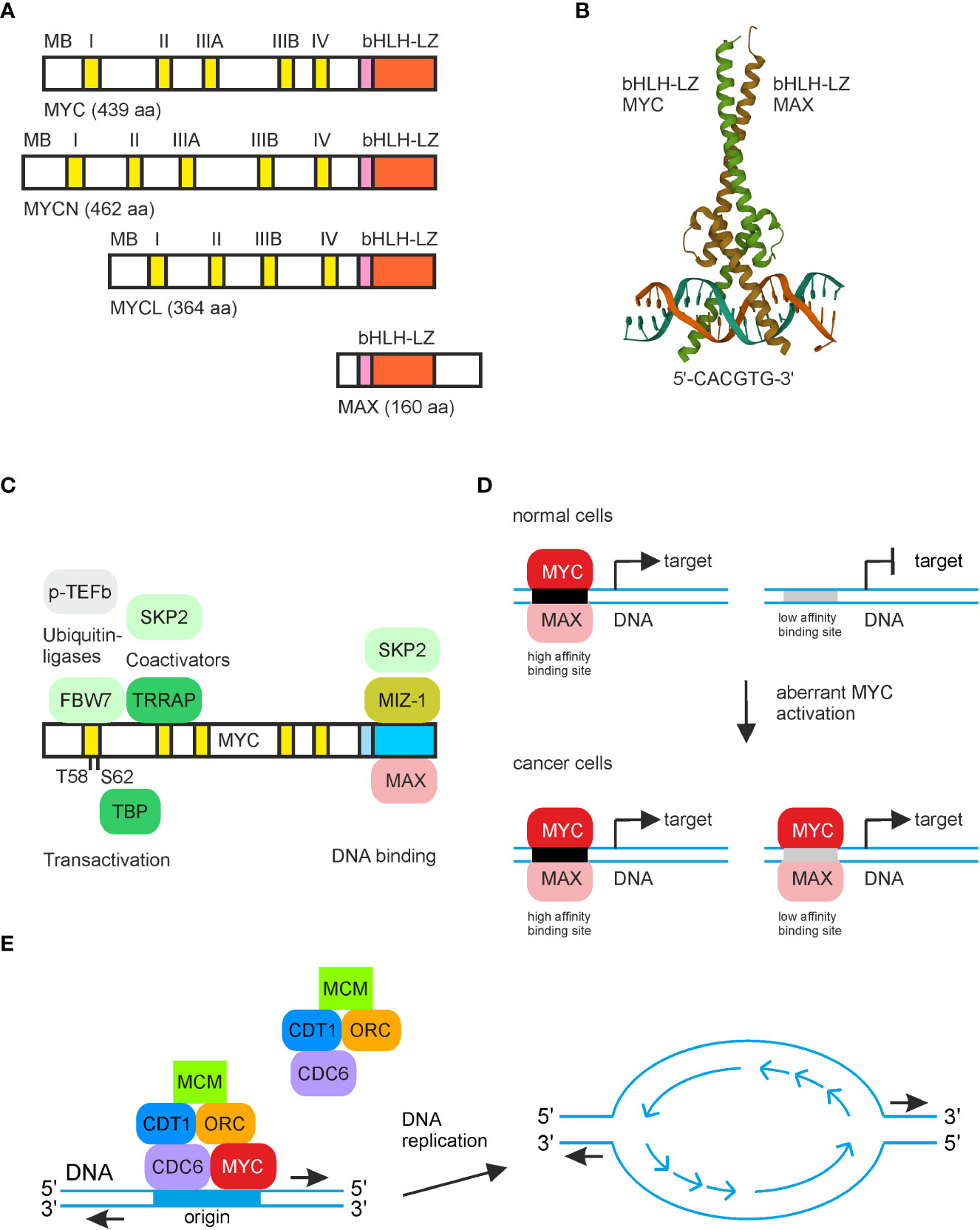
Intrinsically disordered regions in the transcription factor MYC:MAX modulate DNA binding via intramolecular interactions | bioRxiv

Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA | Biochemistry

Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. - Abstract - Europe PMC
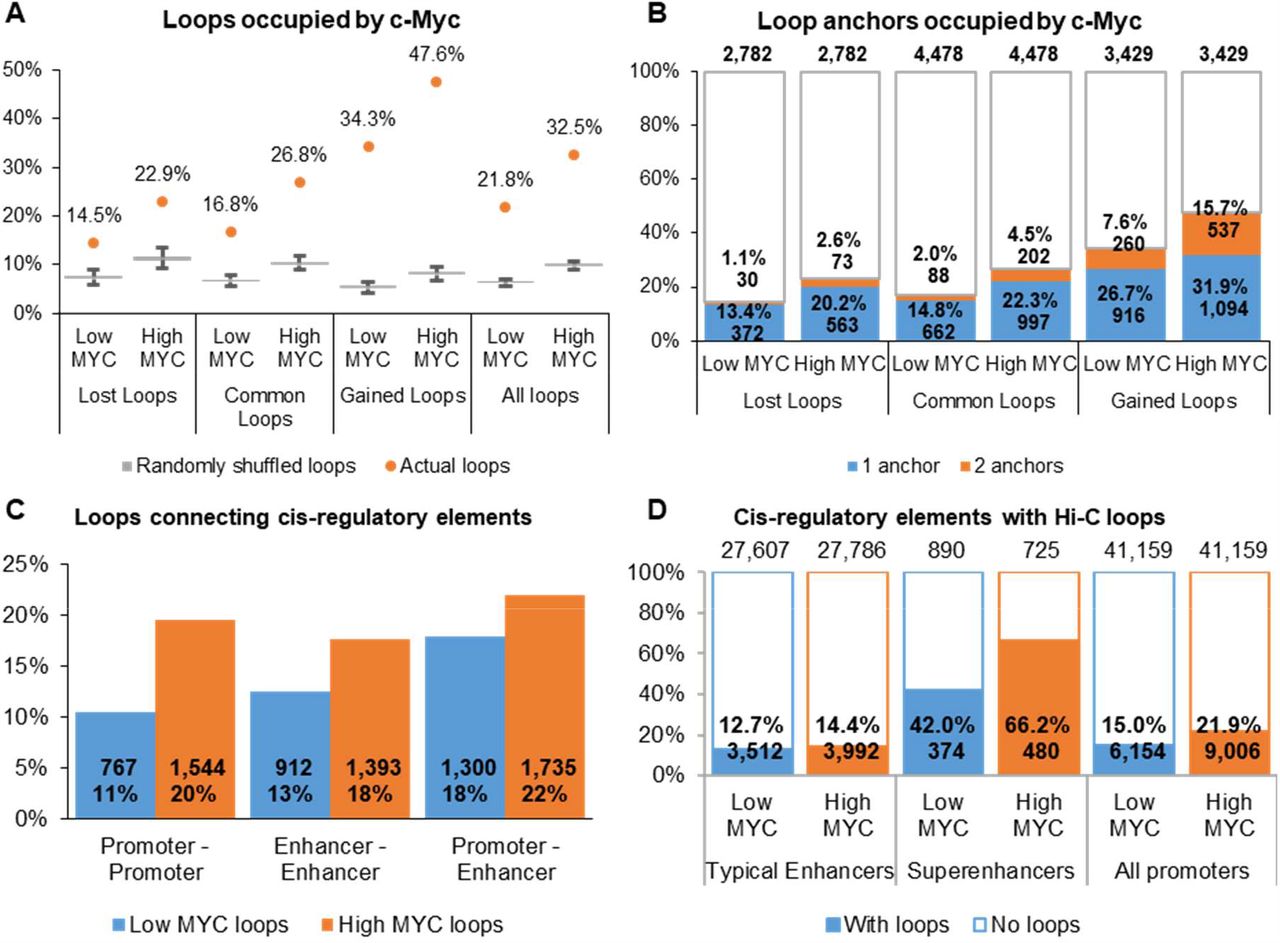
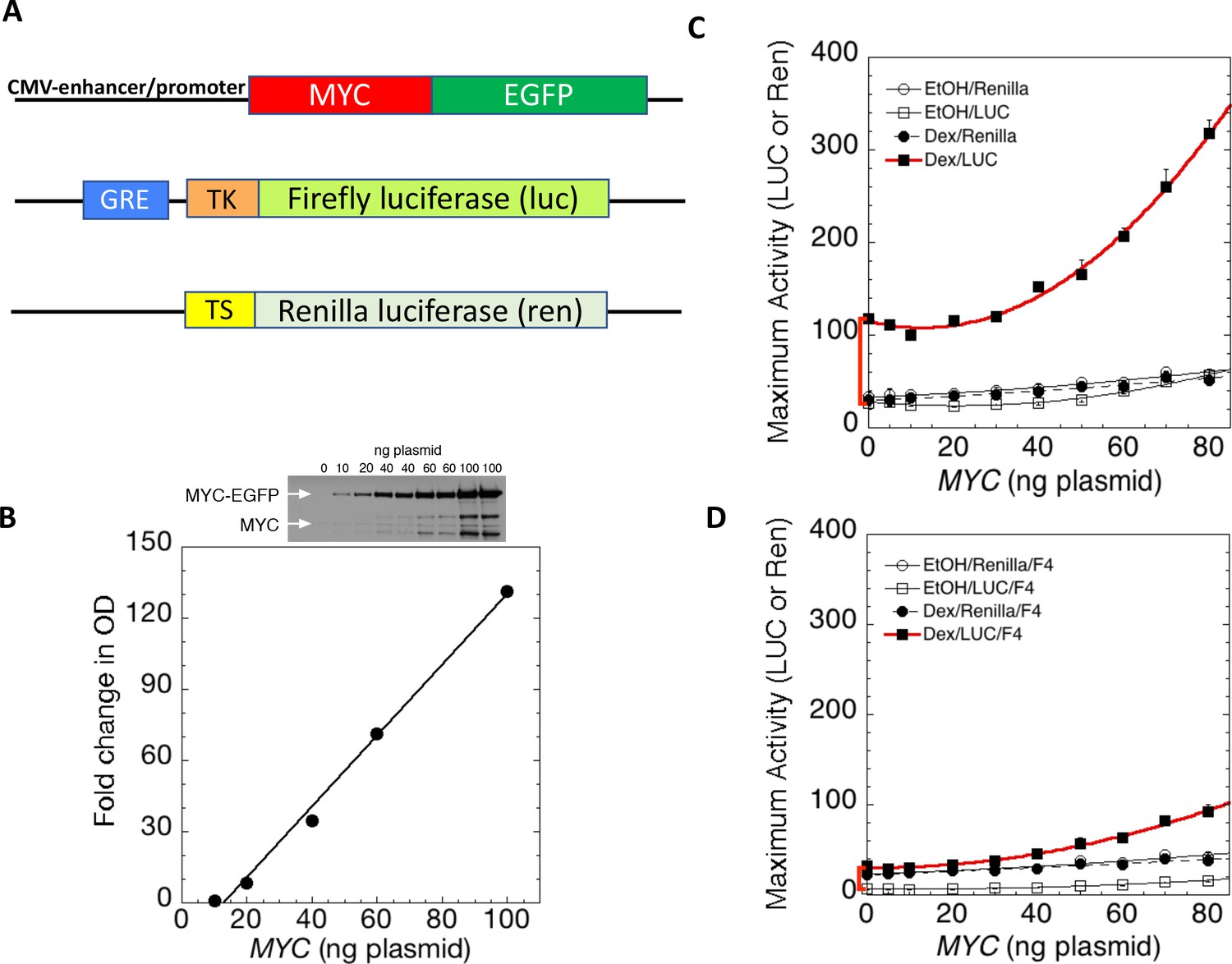
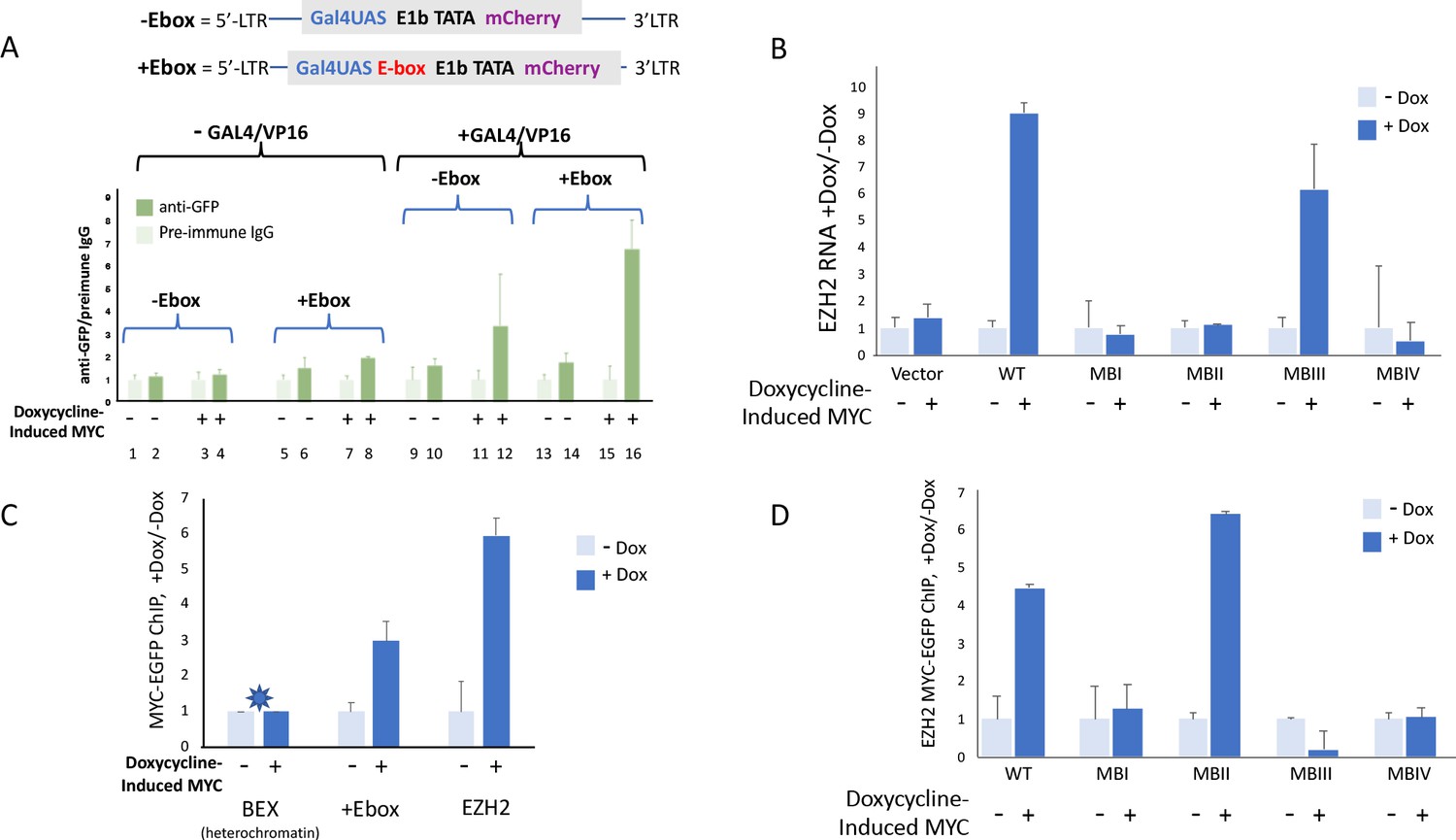
MYC overexpression leads to increased chromatin interactions at superenhancers and c-Myc binding sites | bioRxiv

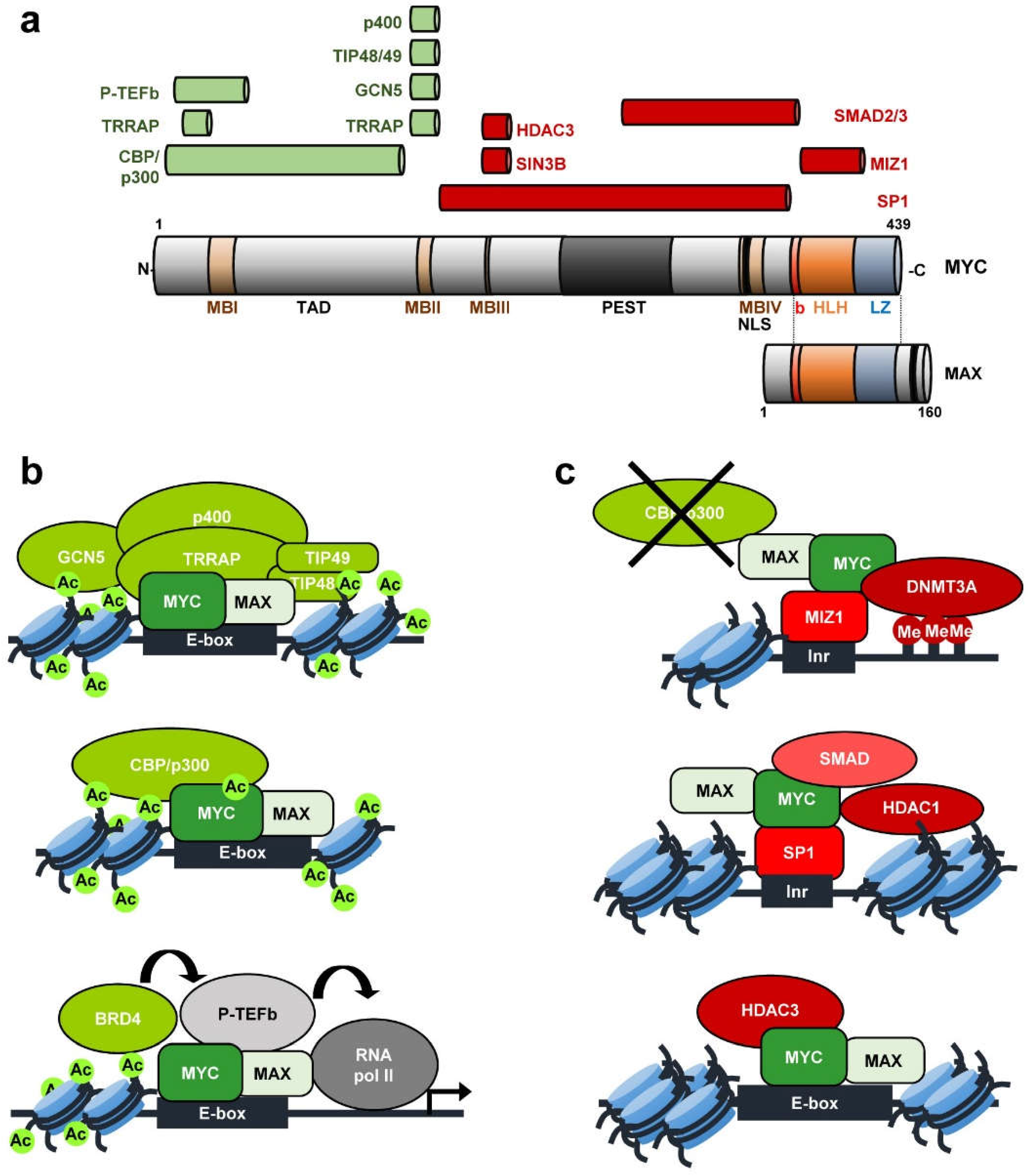
Interaction of the oncoprotein transcription factor MYC with its chromatin cofactor WDR5 is essential for tumor maintenance | PNAS

footprintDB - a database of transcription factors with annotated cis elements (DNA motifs and sites) and binding interfaces
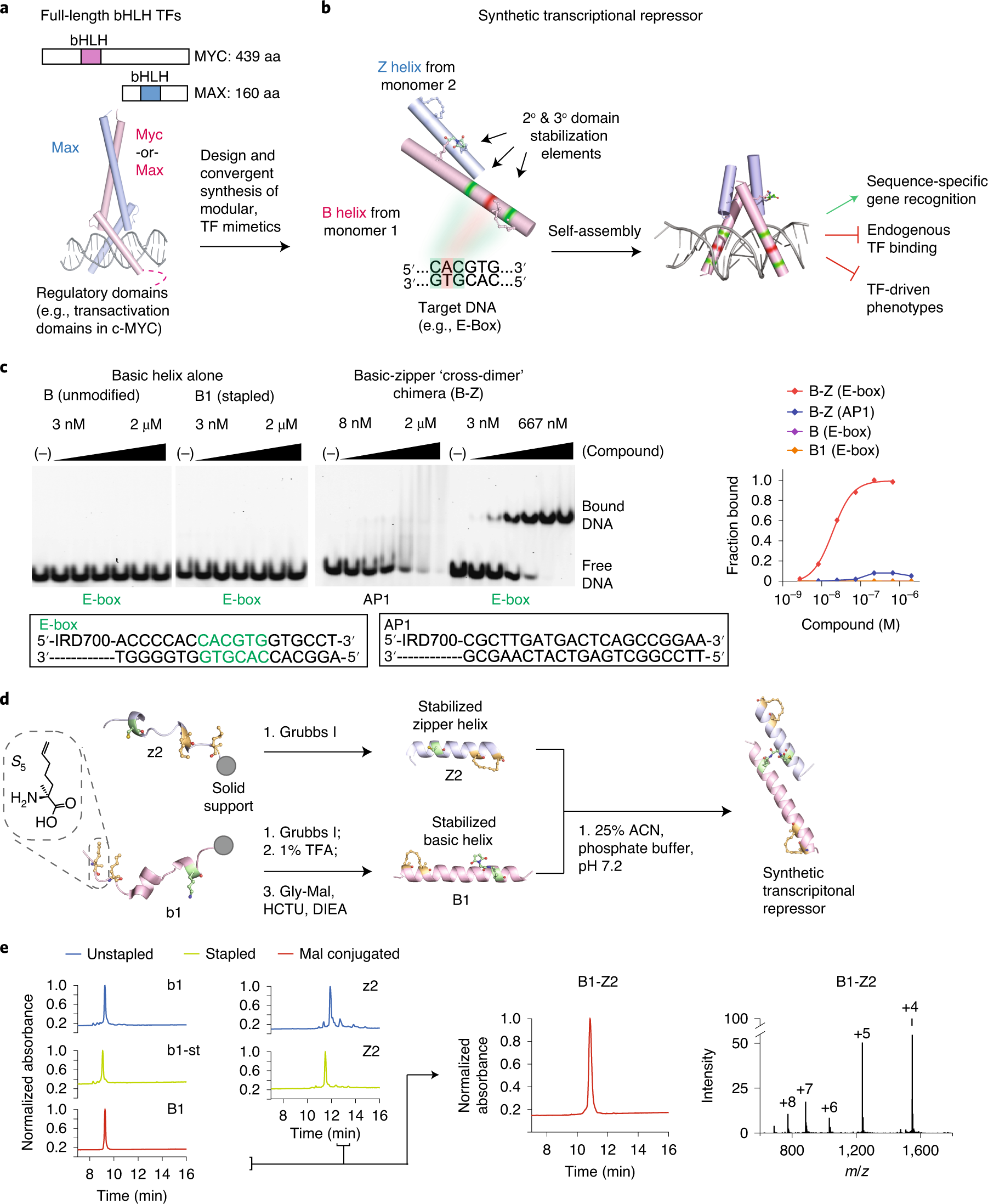
Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains | Nature Biotechnology
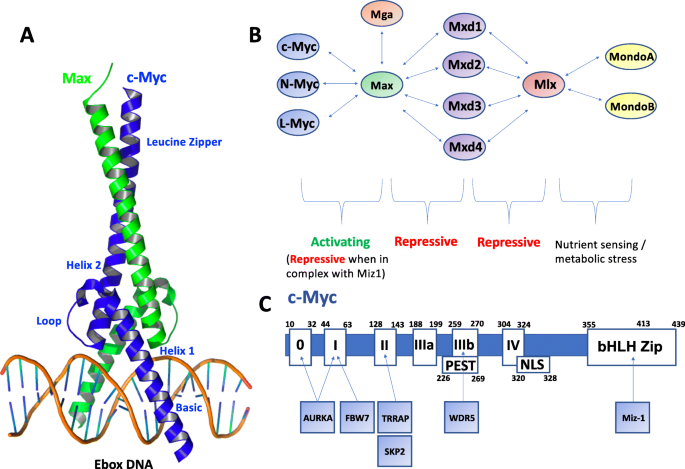
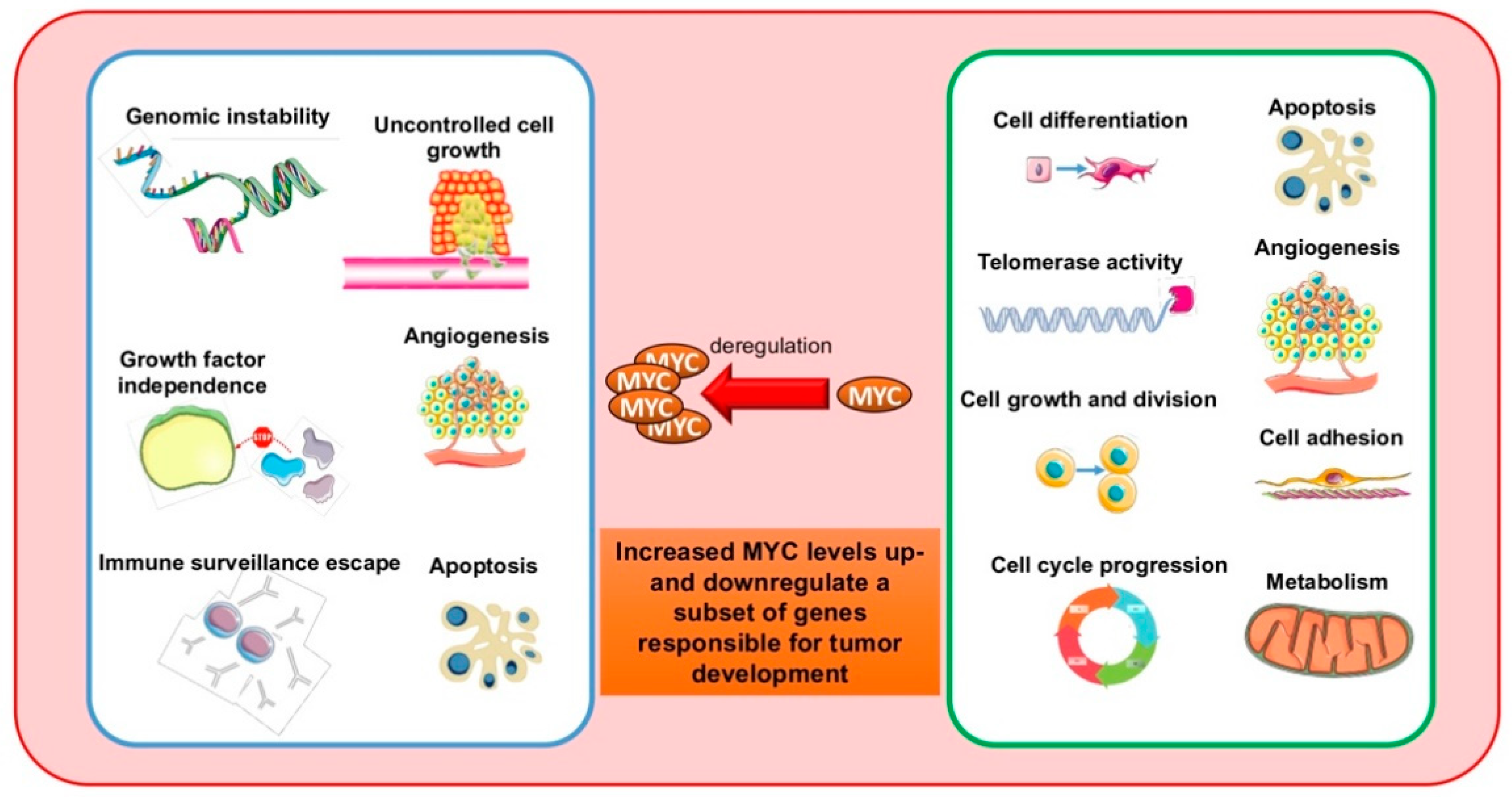
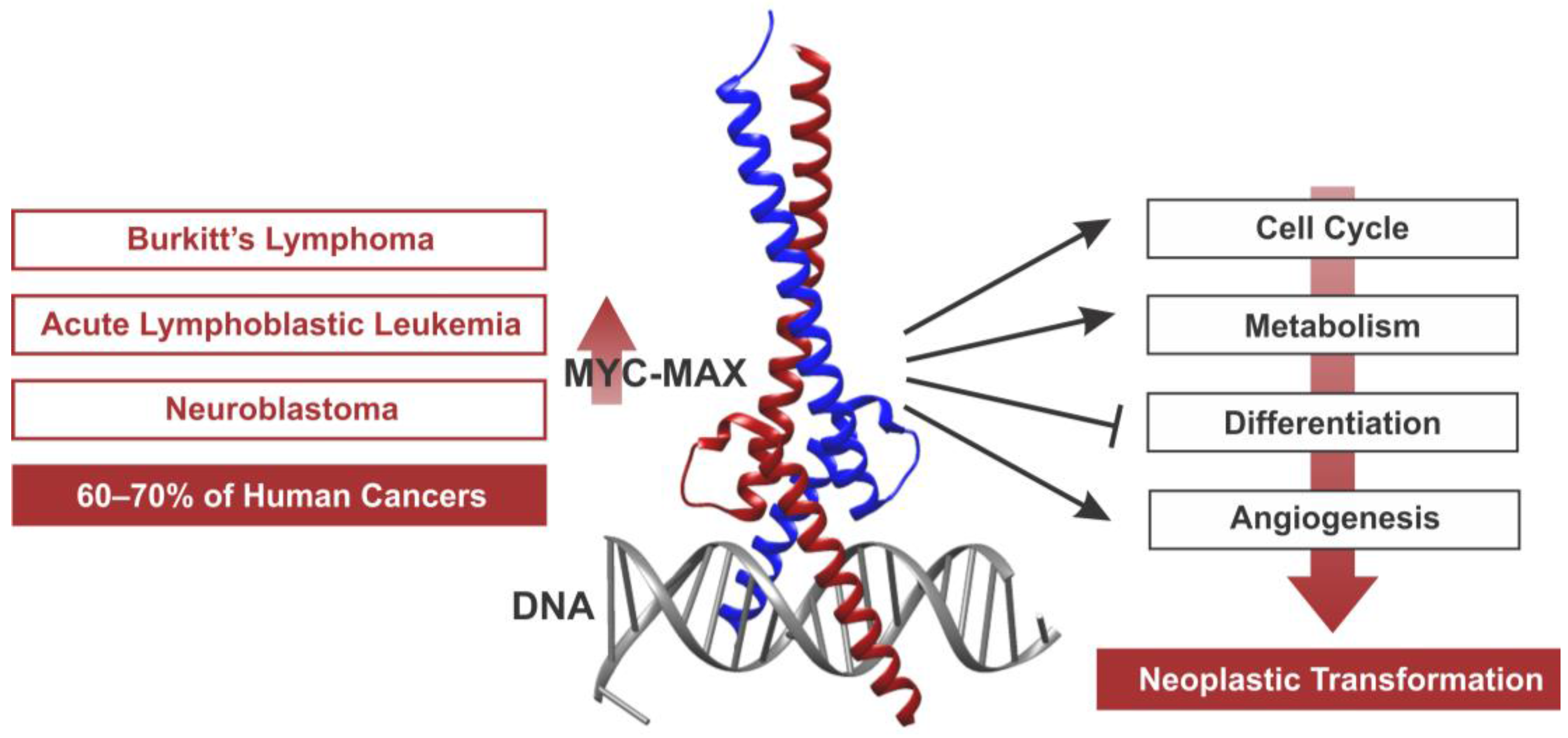
Taking the Myc out of cancer: toward therapeutic strategies to directly inhibit c-Myc | Molecular Cancer | Full Text