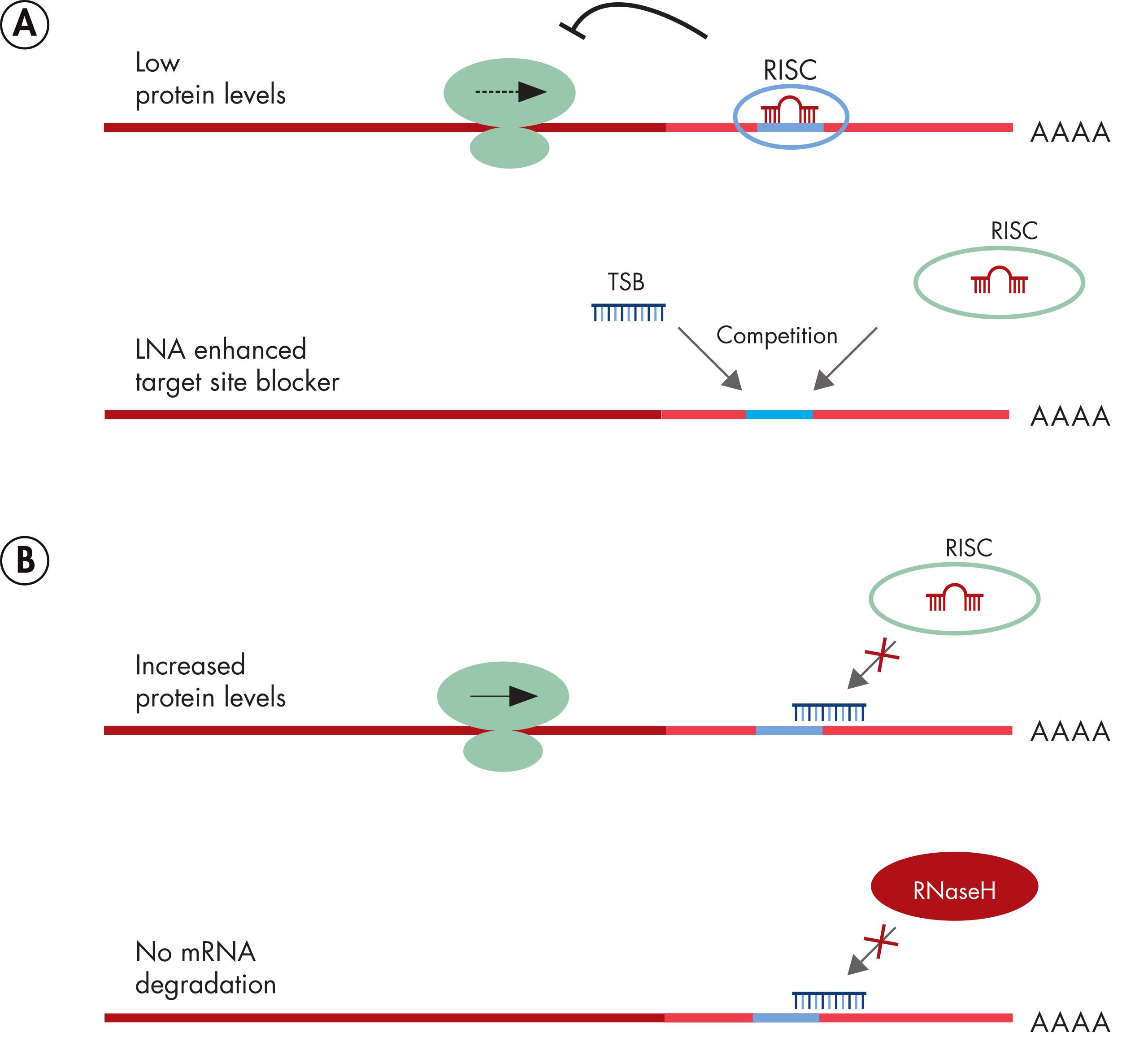
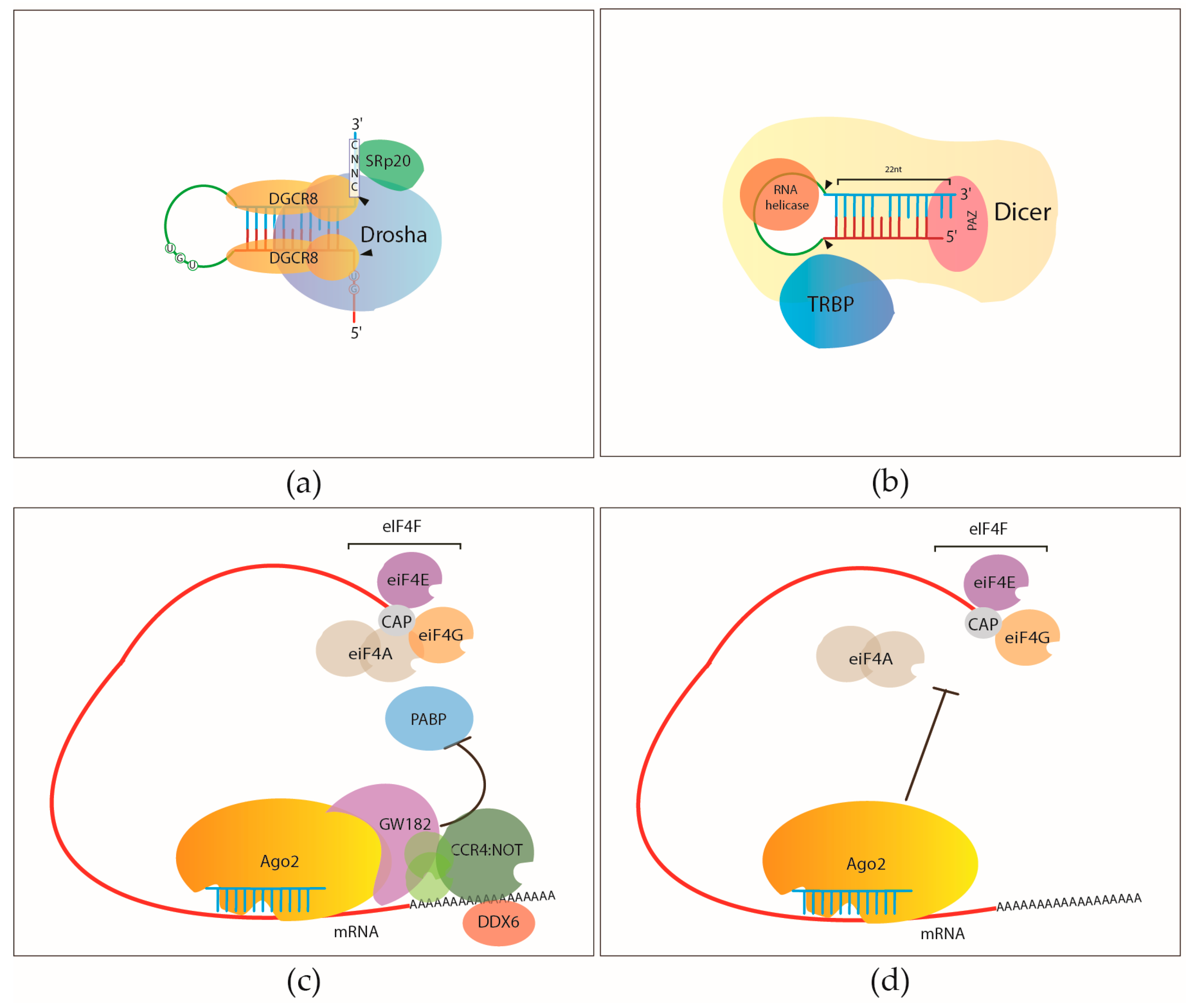
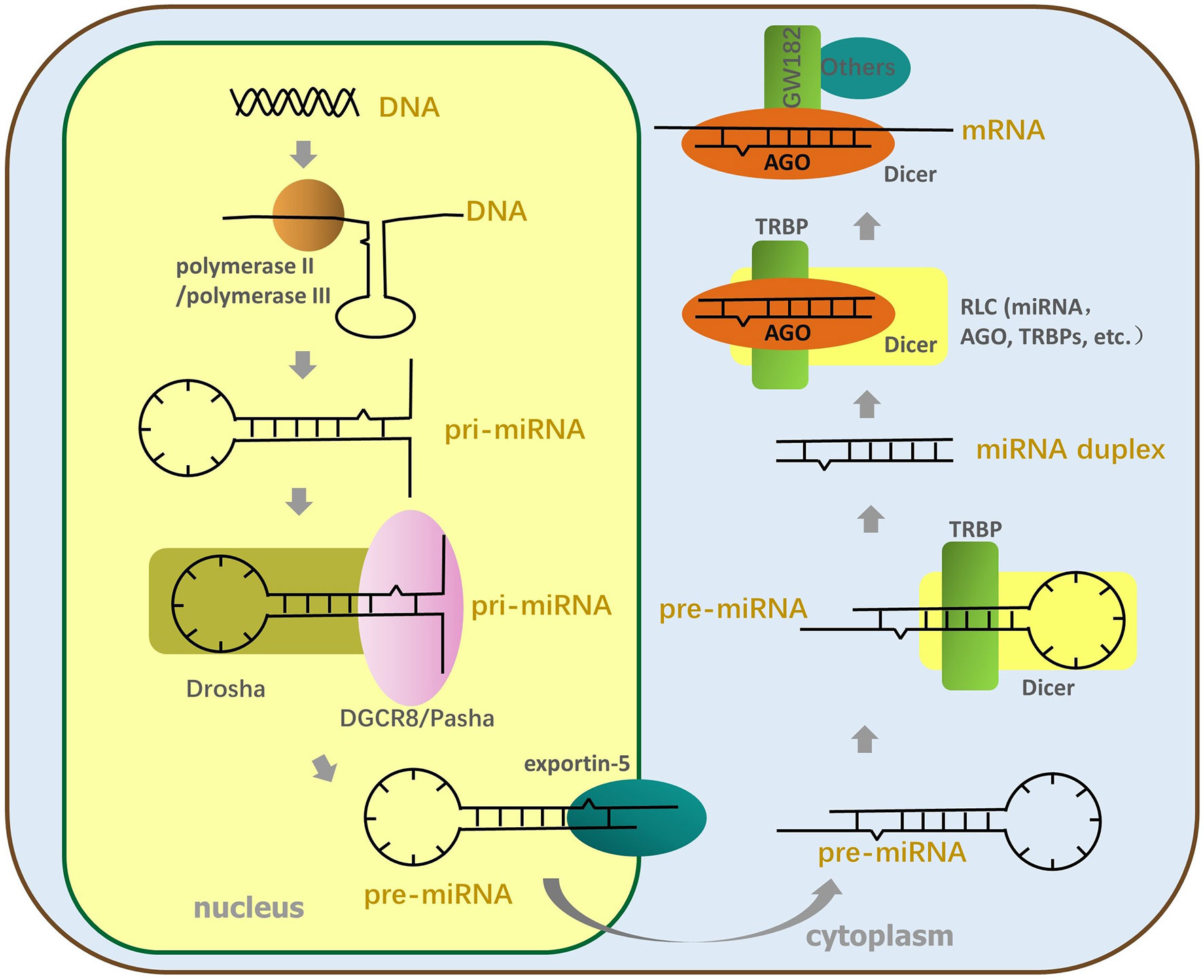
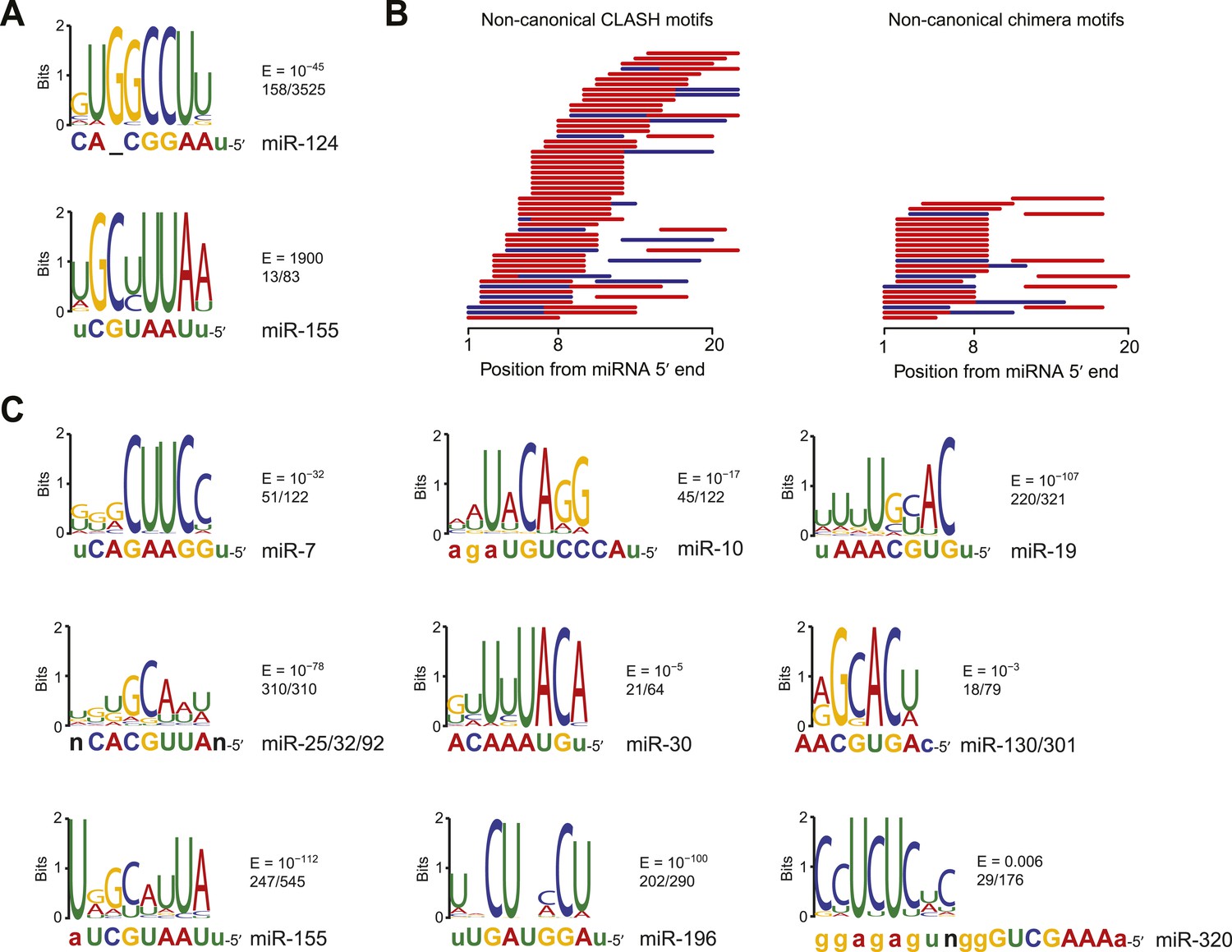
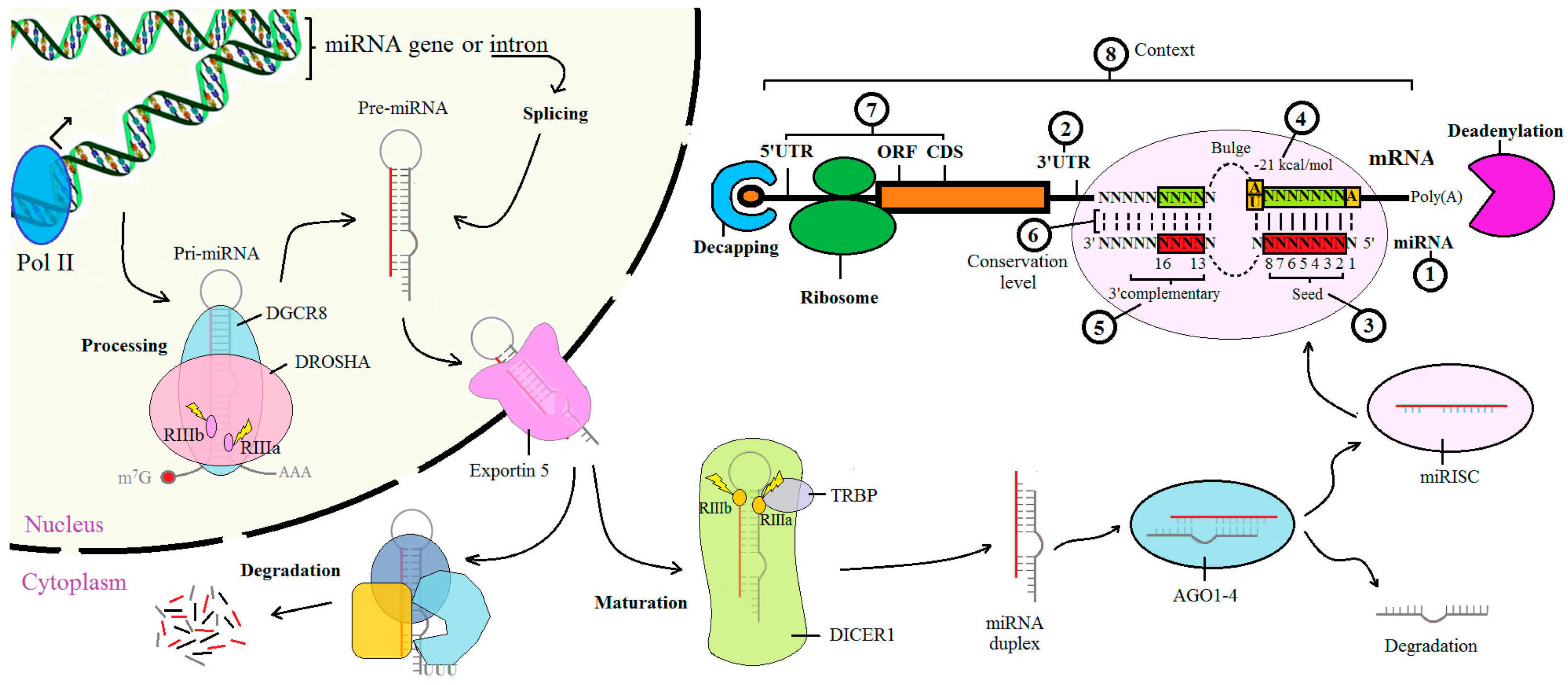
Prediction of the miRNA interactome – Established methods and upcoming perspectives - Computational and Structural Biotechnology Journal
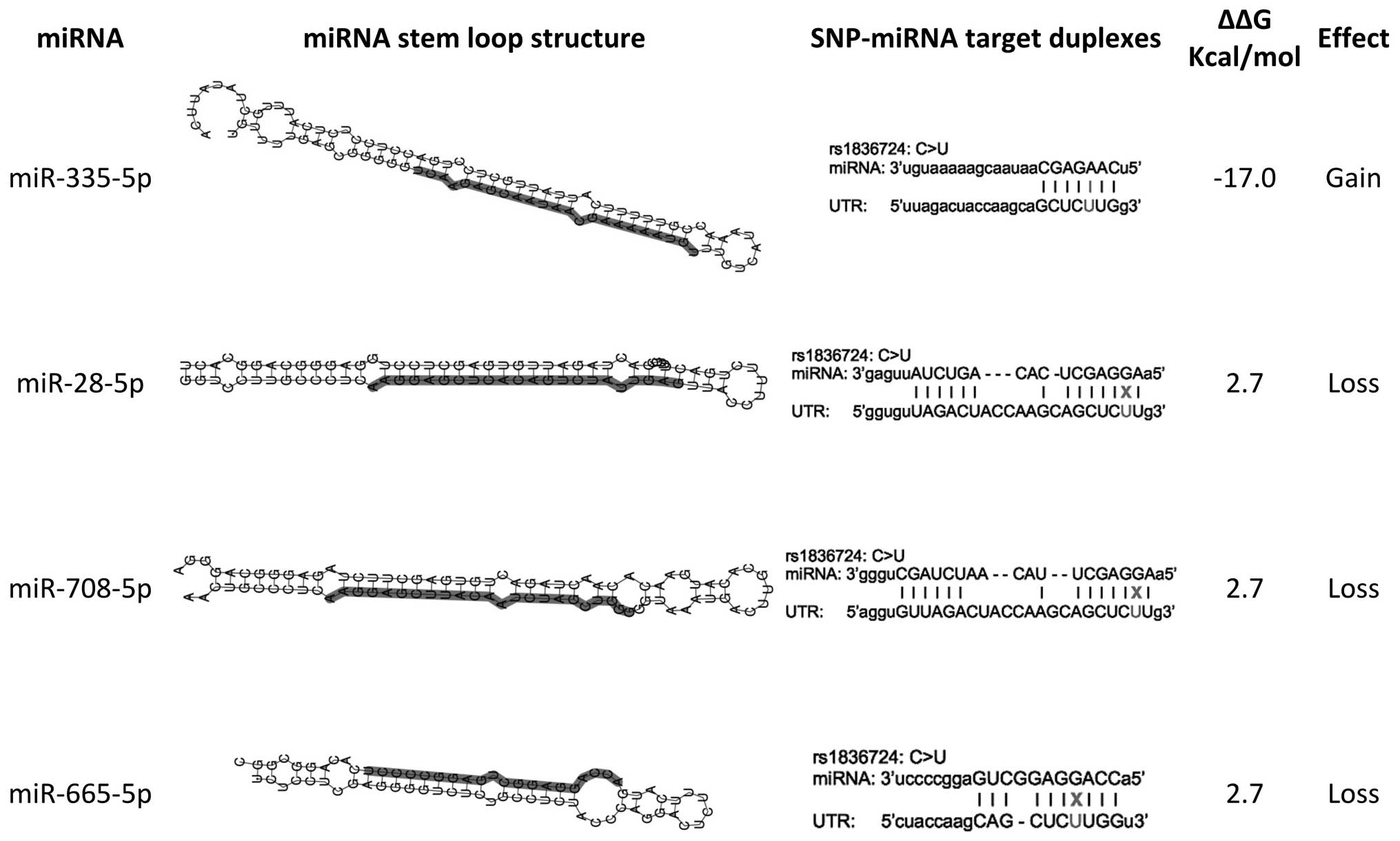
Tumor-promoting function of single nucleotide polymorphism rs1836724 (C3388T) alters multiple potential legitimate microRNA binding sites at the 3'-untranslated region of ErbB4 in breast cancer

Gene silencing by microRNAs: contributions of translational repression and mRNA decay | Nature Reviews Genetics

Mapping of RBP and miRNA binding sites on human 3′UTRs. RBP binding... | Download Scientific Diagram
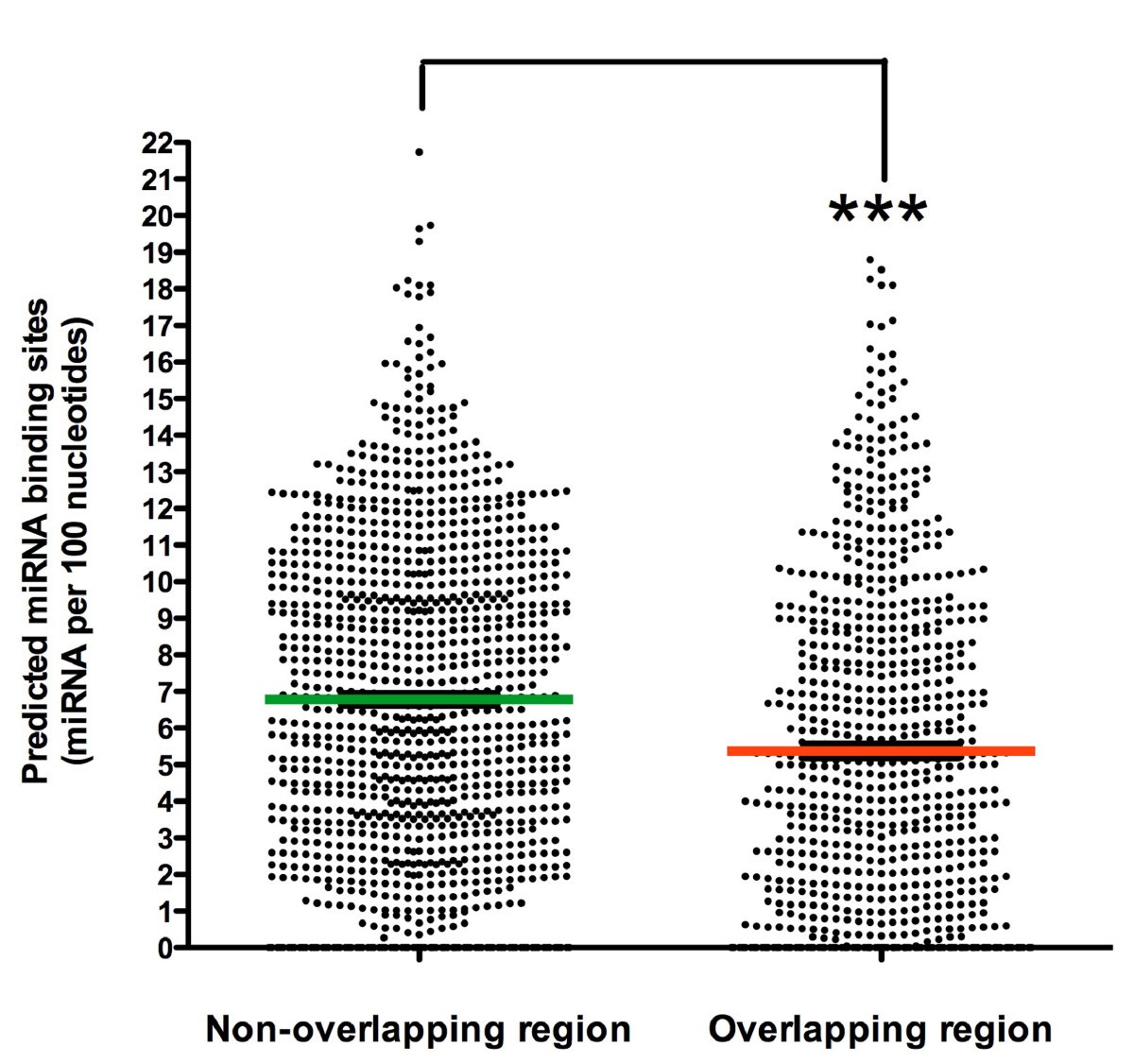
Evidence for natural antisense transcript-mediated inhibition of microRNA function | Genome Biology | Full Text

MicroRNA MIMIC binding sites: Minor flanking nucleotide alterations can strongly impact MIMIC silencing efficacy in Arabidopsis - Wong - 2018 - Plant Direct - Wiley Online Library

Predicted miRNA binding sites in the IL22RA2 3 ′ UTR possibly affected... | Download Scientific Diagram

Canonical and non-canonical miRNA target site types. (A) Diagram of the... | Download Scientific Diagram

The impact of non-synonymous mutations on miRNA binding sites within the SARS-CoV-2 NSP3 and NSP4 genes | Scientific Reports

Identification and consequences of miRNA–target interactions — beyond repression of gene expression | Nature Reviews Genetics

Schema of miRNA binding sites in corresponding 3′-UTR sequence of chicken predicated target gene (seed sequence highlighted in red).