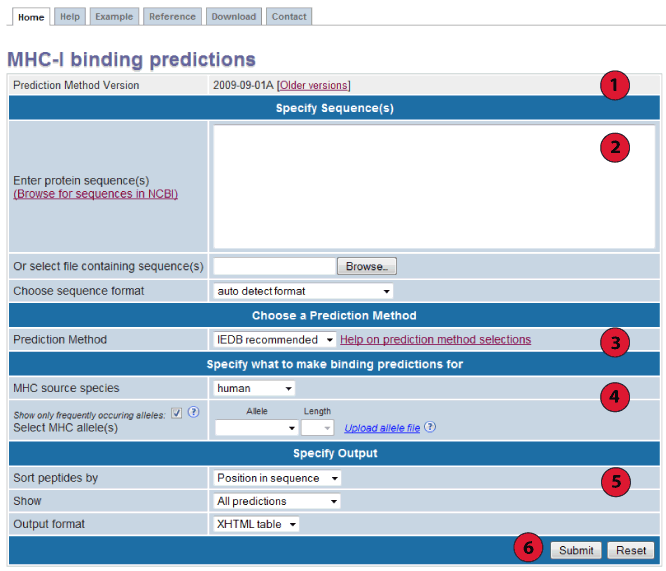
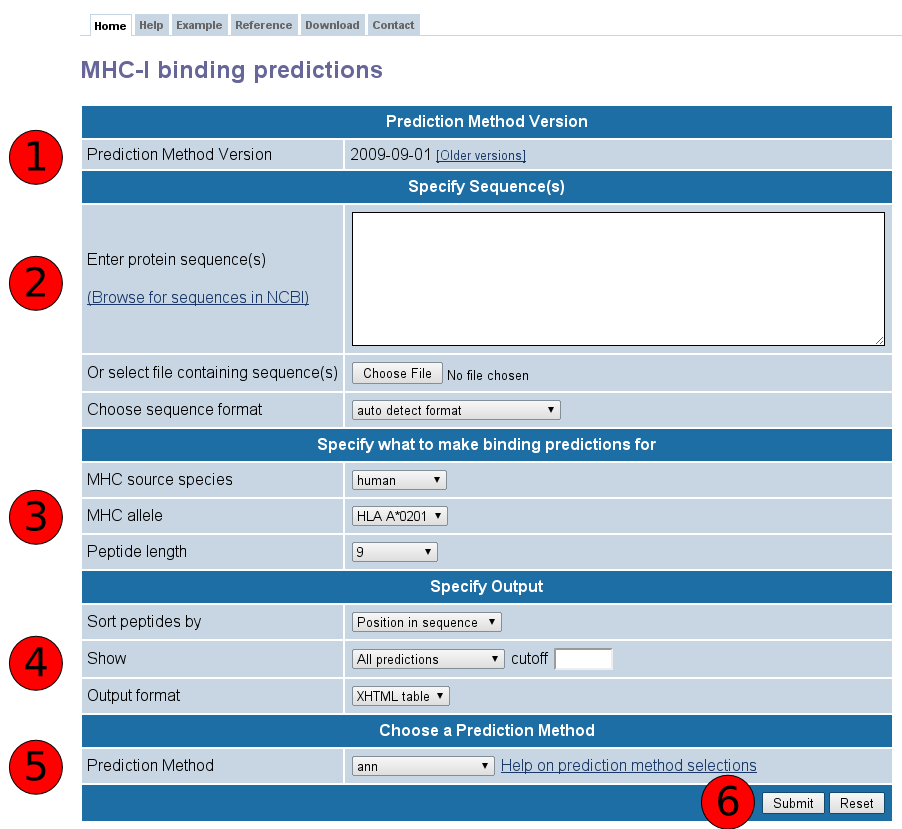
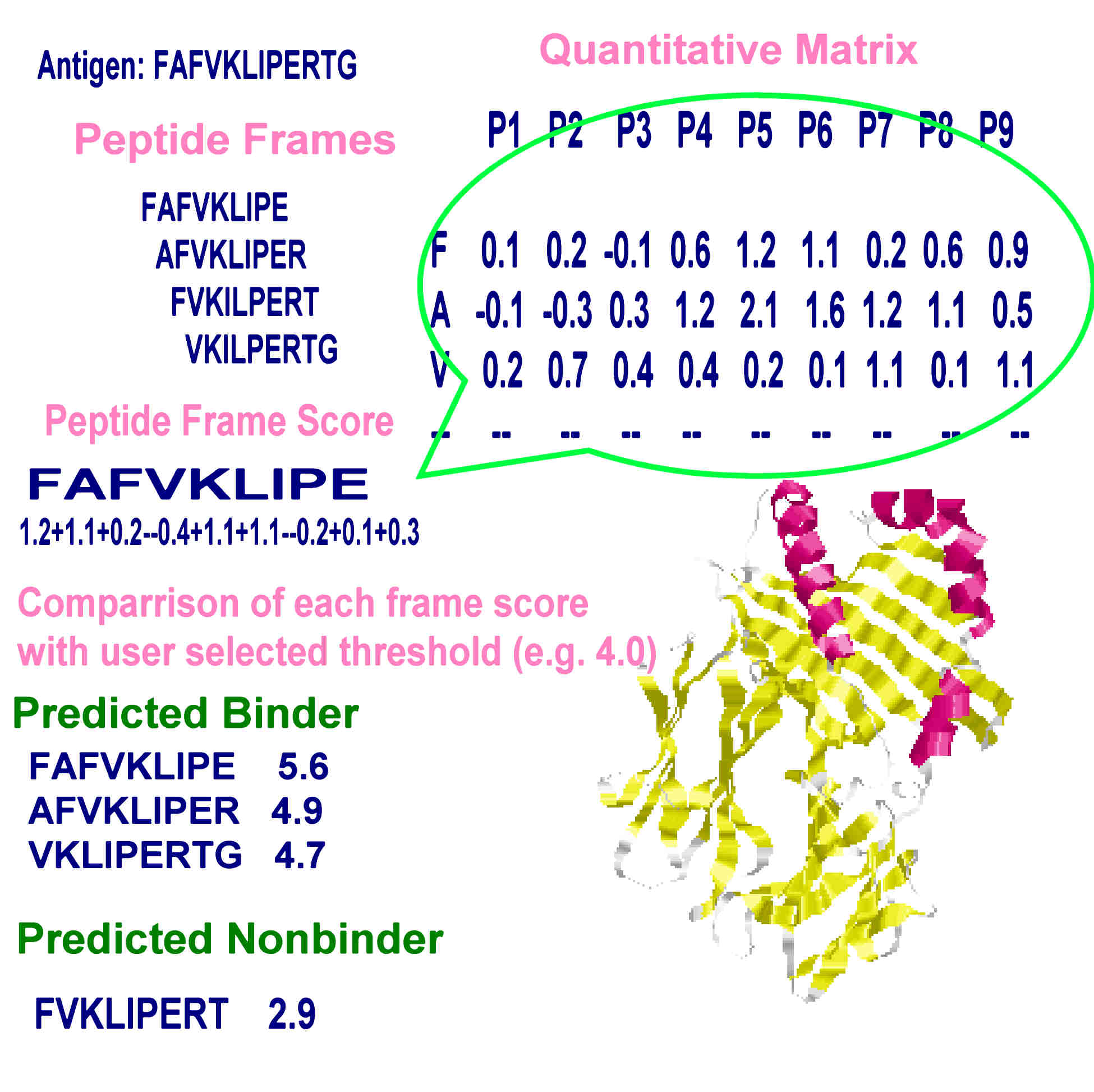
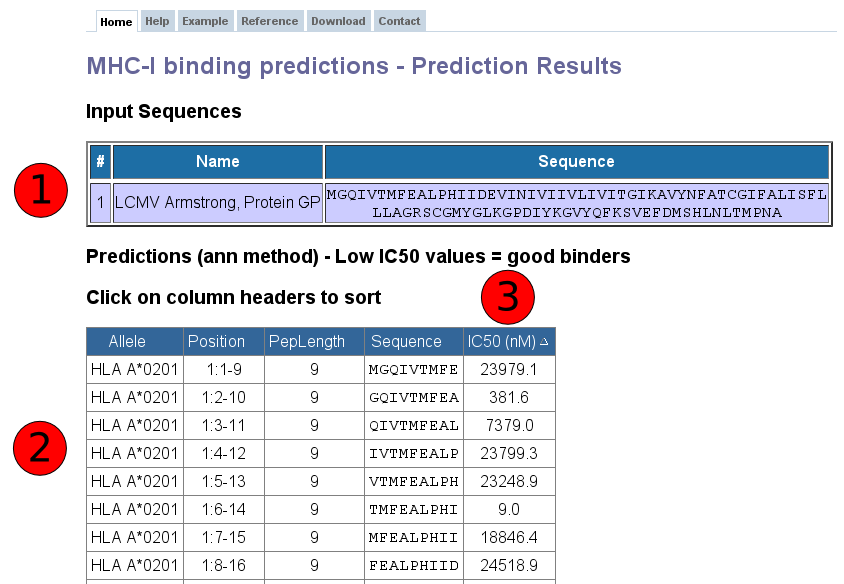
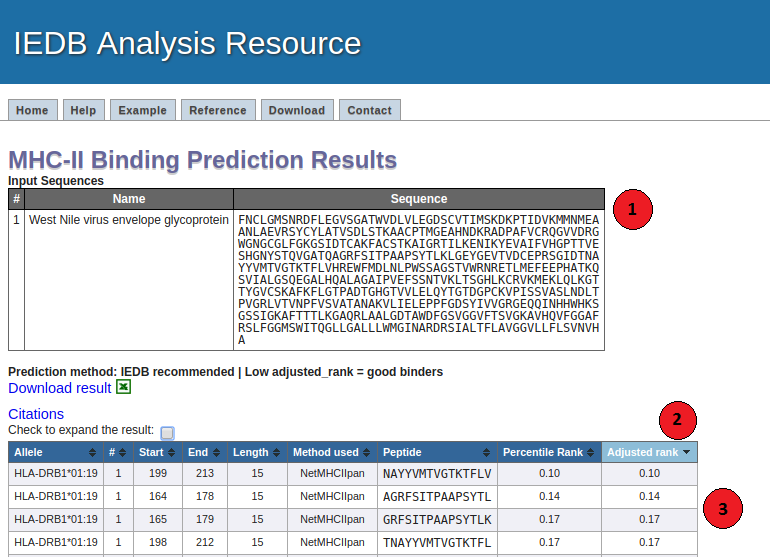
Towardmore accurate pan-specific MHC-peptide binding prediction : a review of current methods and tools | Semantic Scholar

Multiclass classification performance of MHC class I binding epitope... | Download Scientific Diagram
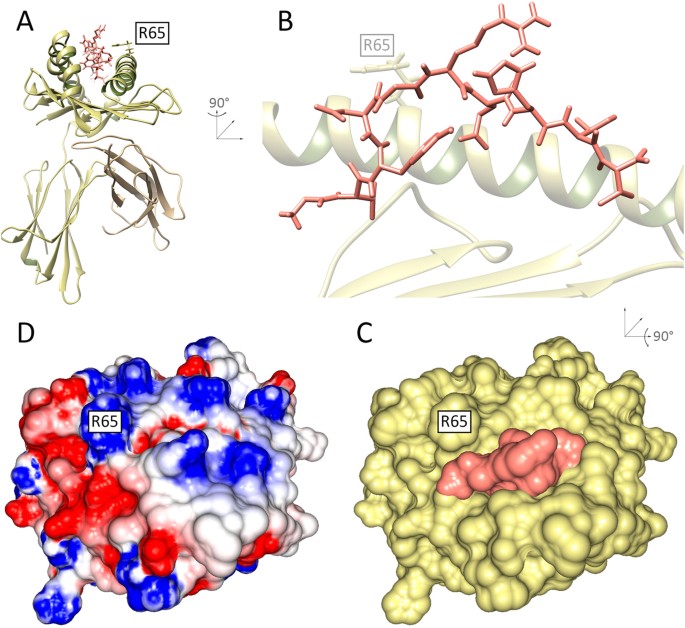
General Prediction of Peptide-MHC Binding Modes Using Incremental Docking: A Proof of Concept | Scientific Reports
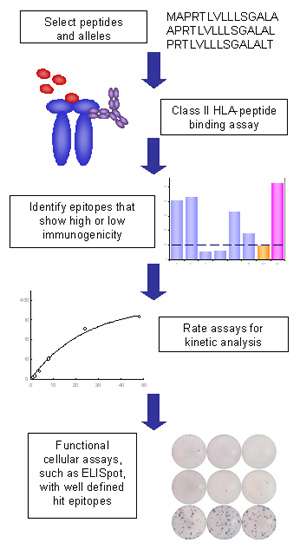
MHC Class II Binding Assays - ProImmune - Mastering Immunity _ MHC pentamers, CD1d tetramers, custom peptide synthesis, immunoassays, T cell epitope discovery, HLA tissue typing, cellular analysis, ligand binding assays, immune response

Structure-based Methods for Binding Mode and Binding Affinity Prediction for Peptide-MHC Complexes | Bentham Science
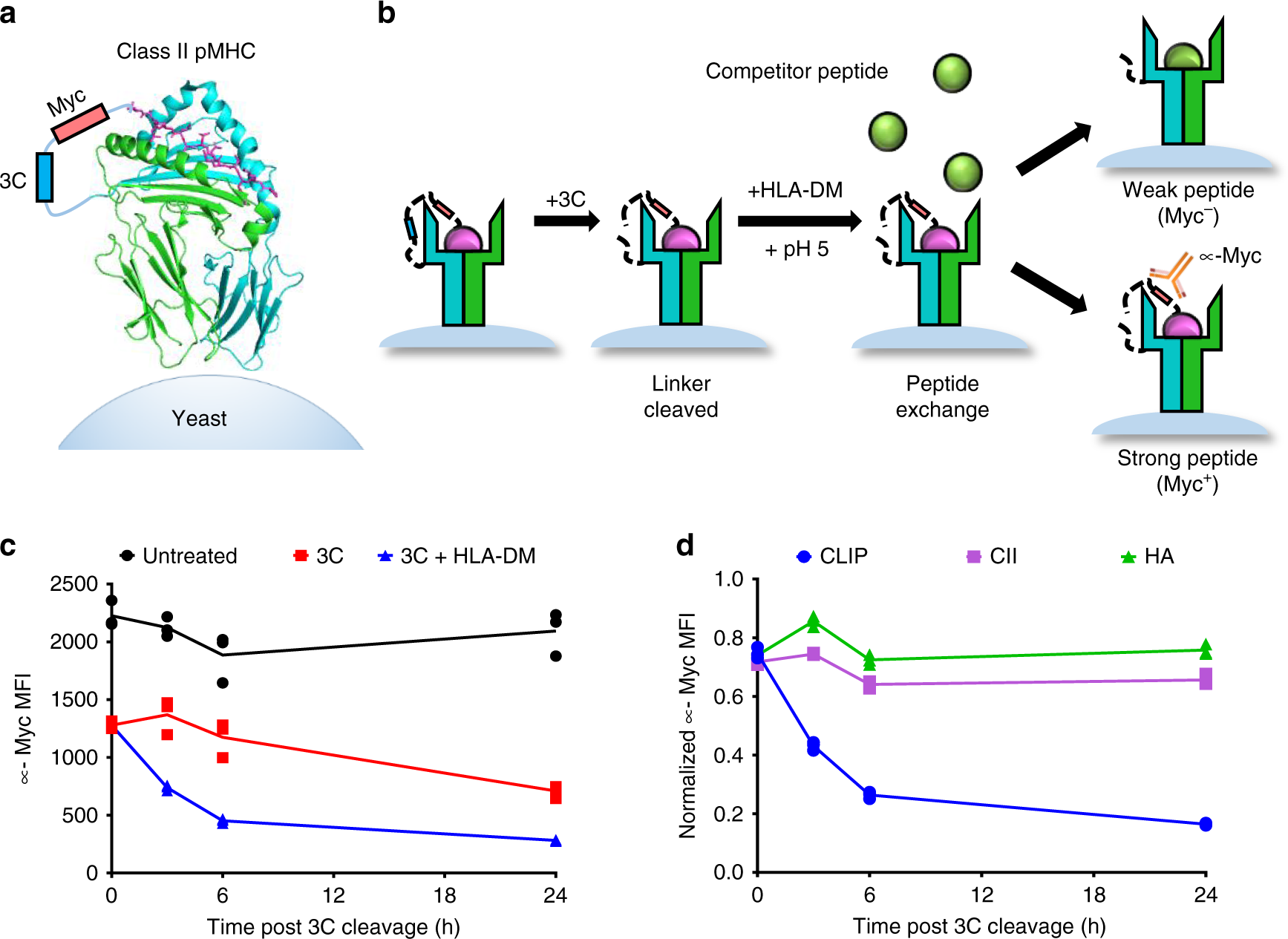
Repertoire-scale determination of class II MHC peptide binding via yeast display improves antigen prediction | Nature Communications

Repertoire-scale determination of class II MHC peptide binding via yeast display improves antigen prediction | Nature Communications

Improved methods for predicting peptide binding affinity to MHC class II molecules - Jensen - 2018 - Immunology - Wiley Online Library
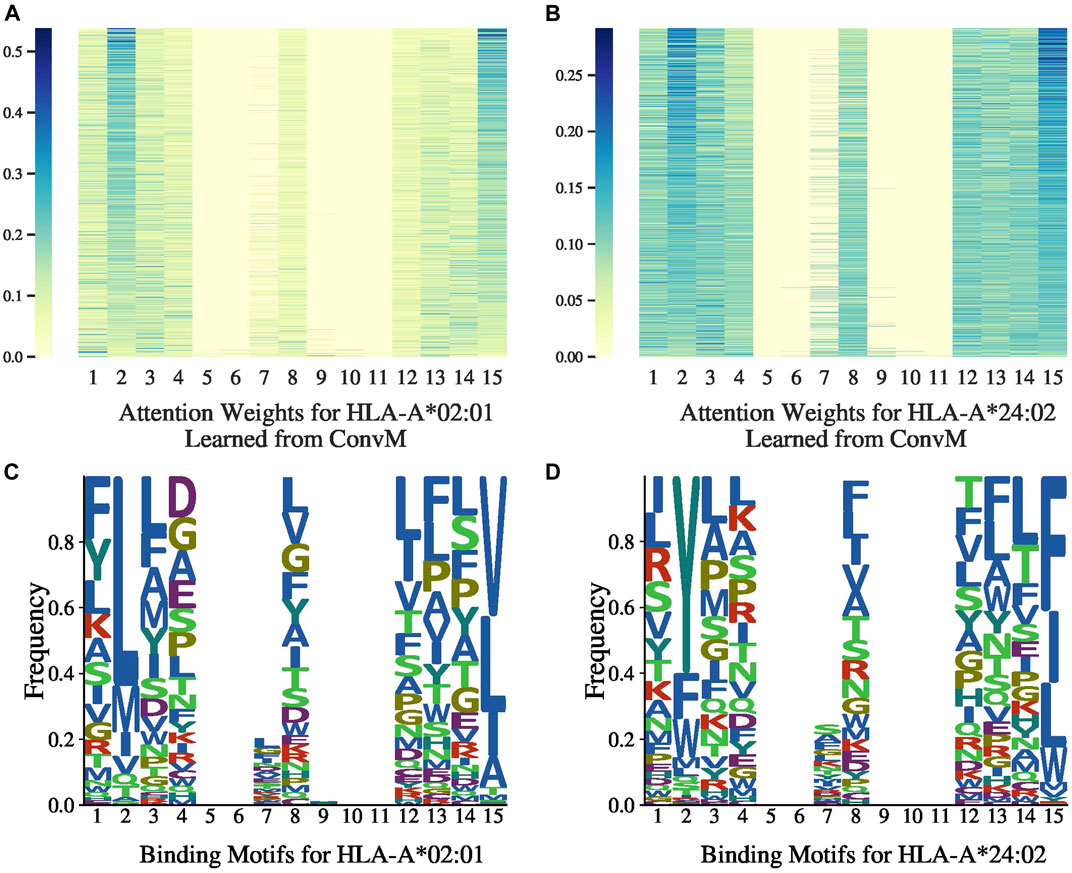
Frontiers | Ranking-Based Convolutional Neural Network Models for Peptide- MHC Class I Binding Prediction

Deciphering the Structural Enigma of HLA Class-II Binding Peptides for Enhanced Immunoinformatics-based Prediction of Vaccine Epitopes | Journal of Proteome Research

Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes - ScienceDirect

NNAlign_MA; MHC Peptidome Deconvolution for Accurate MHC Binding Motif Characterization and Improved T-cell Epitope Predictions - ScienceDirect

Biology | Free Full-Text | Prediction of Major Histocompatibility Complex Binding with Bilateral and Variable Long Short Term Memory Networks