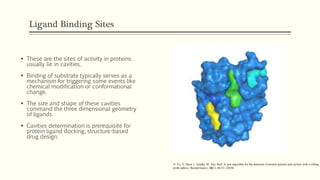
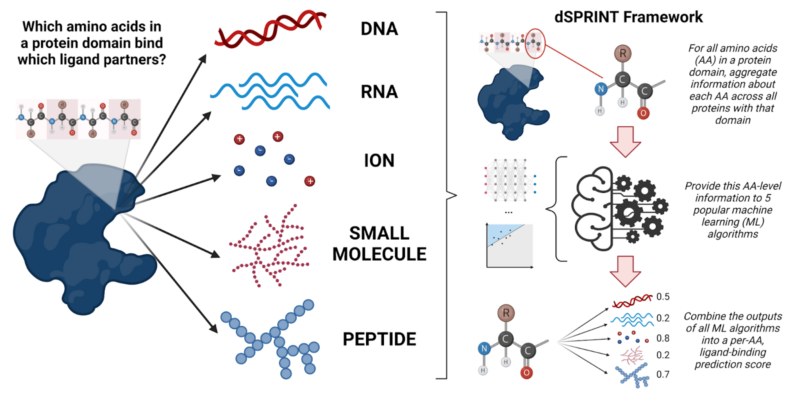
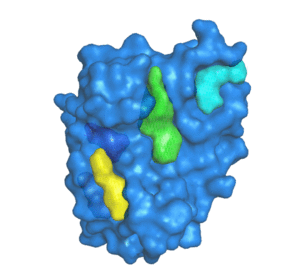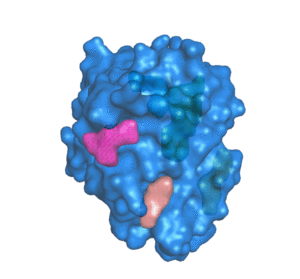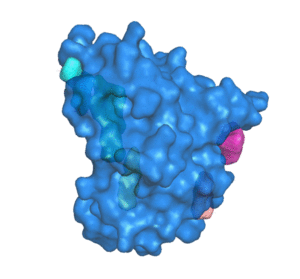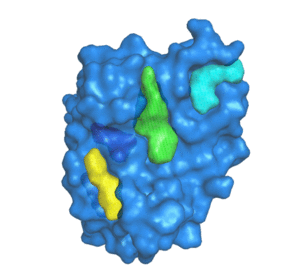
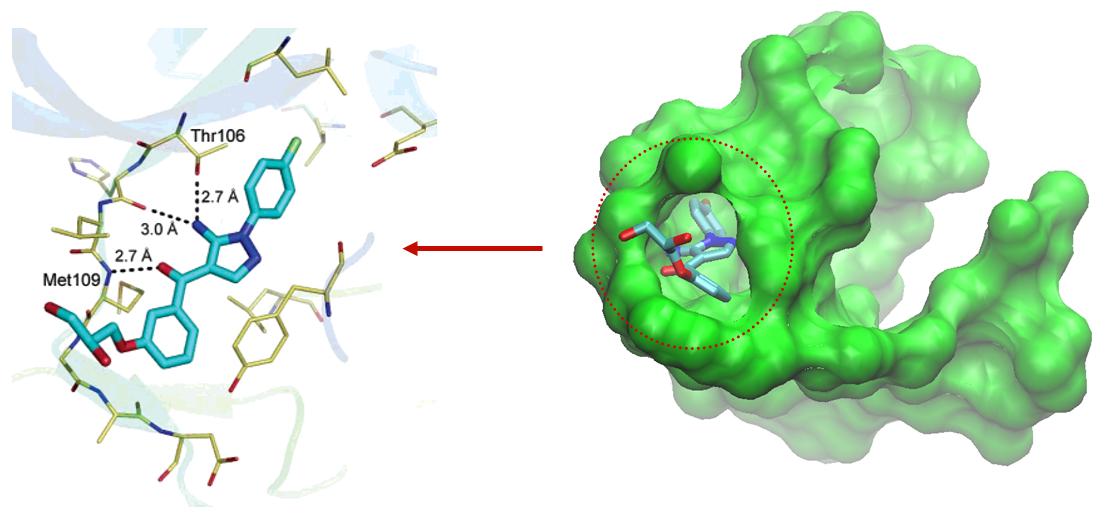
Predicting the impacts of mutations on protein-ligand binding affinity based on molecular dynamics simulations and machine learning methods - Computational and Structural Biotechnology Journal
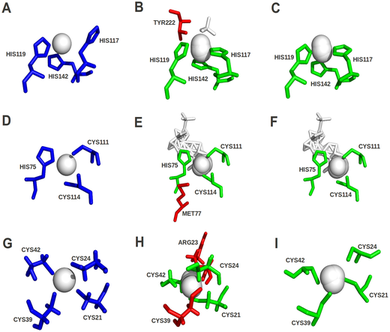
IJMS | Free Full-Text | Proteins and Their Interacting Partners: An Introduction to Protein–Ligand Binding Site Prediction Methods

Workflow of binding sites detection. Each protein chain submitted would... | Download Scientific Diagram

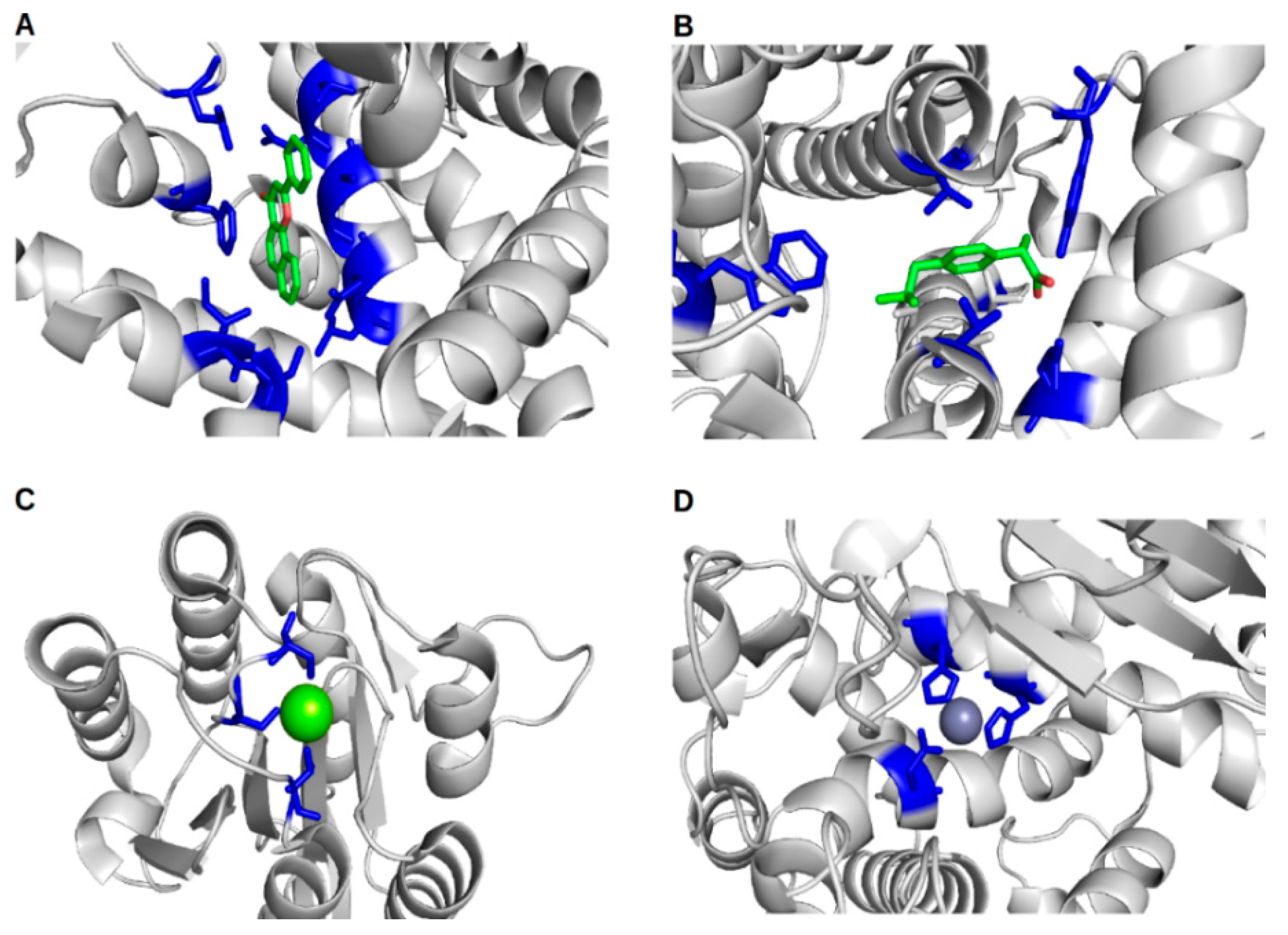
Can We Rely on Computational Predictions To Correctly Identify Ligand Binding Sites on Novel Protein Drug Targets? Assessment of Binding Site Prediction Methods and a Protocol for Validation of Predicted Binding Sites

Protein-ligand binding affinity prediction based on profiles of intermolecular contacts - ScienceDirect

PUResNet: prediction of protein-ligand binding sites using deep residual neural network | Journal of Cheminformatics | Full Text

A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation | PNAS
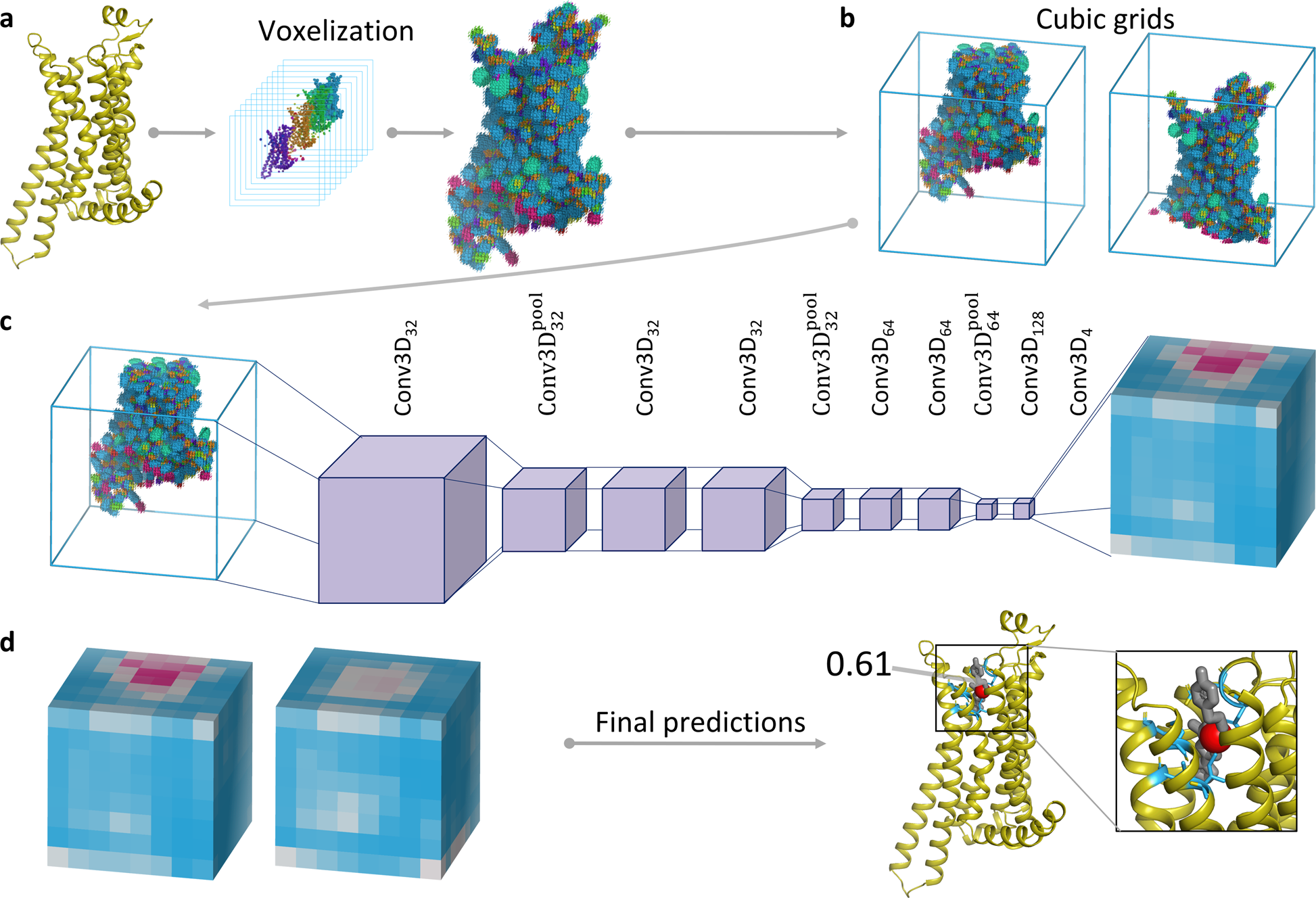
Spatiotemporal identification of druggable binding sites using deep learning | Communications Biology

bSiteFinder, an improved protein-binding sites prediction server based on structural alignment: more accurate and less time-consuming | Journal of Cheminformatics | Full Text

Illustration of protein binding site prediction for Targets T0570 (a),... | Download Scientific Diagram

PUResNet: prediction of protein-ligand binding sites using deep residual neural network | Journal of Cheminformatics | Full Text
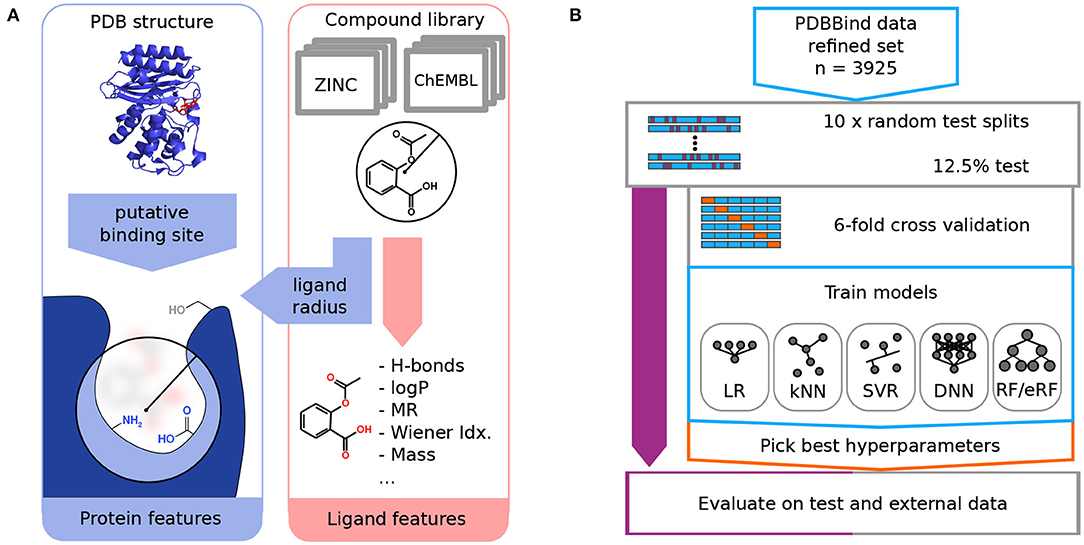
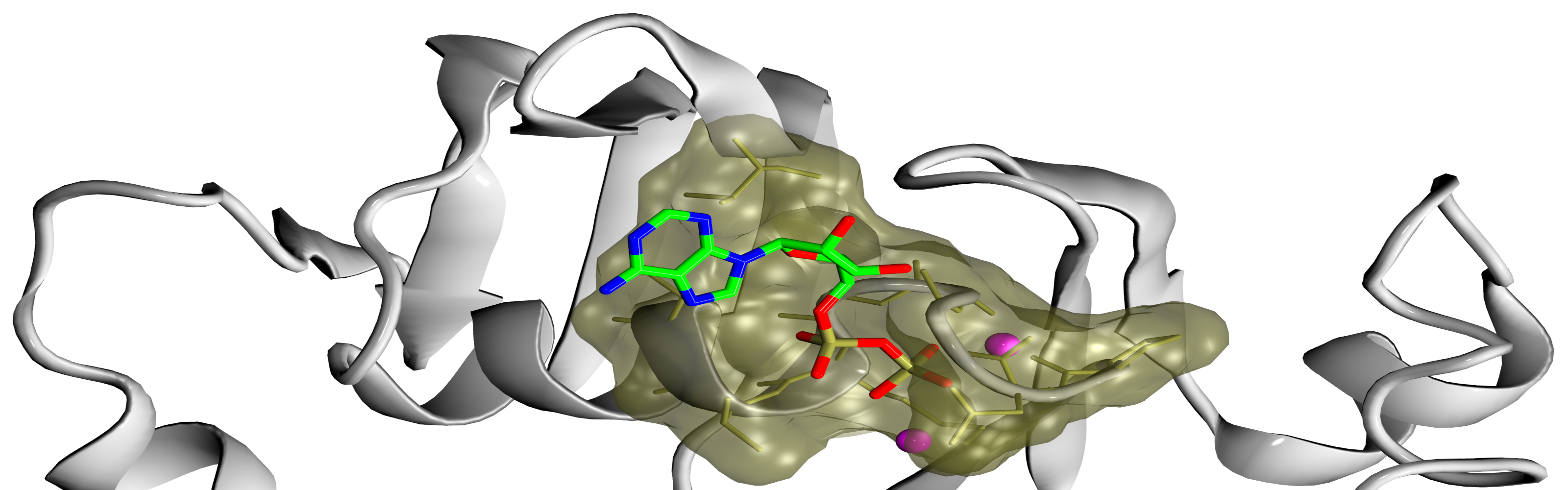
Frontiers | RASPD+: Fast Protein-Ligand Binding Free Energy Prediction Using Simplified Physicochemical Features
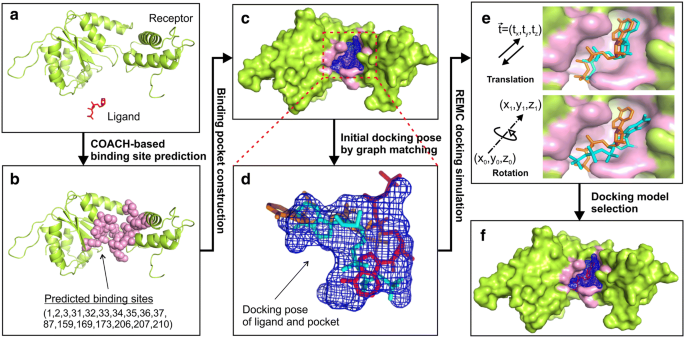
EDock: blind protein–ligand docking by replica-exchange monte carlo simulation | Journal of Cheminformatics | Full Text

Exploring the computational methods for protein-ligand binding site prediction - Computational and Structural Biotechnology Journal

RASPD+: Fast Protein-Ligand Binding Free Energy Prediction Using Simplified Physicochemical Features | Theoretical and Computational Chemistry | ChemRxiv | Cambridge Open Engage

![PDF] The FunFOLD 2 server for the prediction of protein ligand interactions | Semantic Scholar PDF] The FunFOLD 2 server for the prediction of protein ligand interactions | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/c50225f129c18e56d2cbe4352383ce46df326d70/5-Figure1-1.png)