MARIA-Neural network for accurate prediction of HLA class II antigen presentation | Explore Technologies

Defining HLA-II Ligand Processing and Binding Rules with Mass Spectrometry Enhances Cancer Epitope Prediction - ScienceDirect

DeepNetBim: deep learning model for predicting HLA-epitope interactions based on network analysis by harnessing binding and immunogenicity information | BMC Bioinformatics | Full Text

Deciphering the Structural Enigma of HLA Class-II Binding Peptides for Enhanced Immunoinformatics-based Prediction of Vaccine Epitopes | Journal of Proteome Research

Towardmore accurate pan-specific MHC-peptide binding prediction : a review of current methods and tools | Semantic Scholar
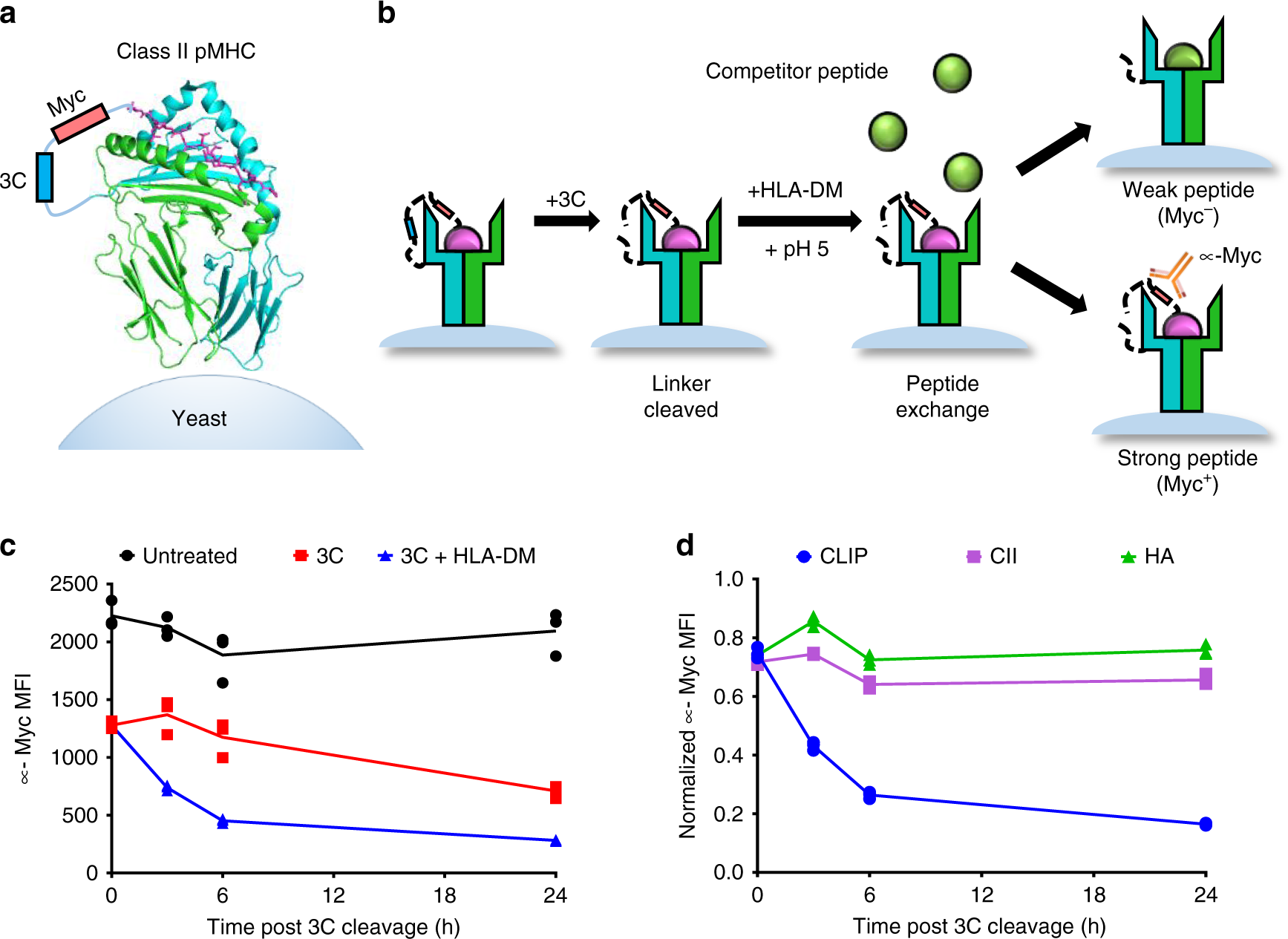
Repertoire-scale determination of class II MHC peptide binding via yeast display improves antigen prediction | Nature Communications

Deciphering the Structural Enigma of HLA Class-II Binding Peptides for Enhanced Immunoinformatics-based Prediction of Vaccine Epitopes | Journal of Proteome Research

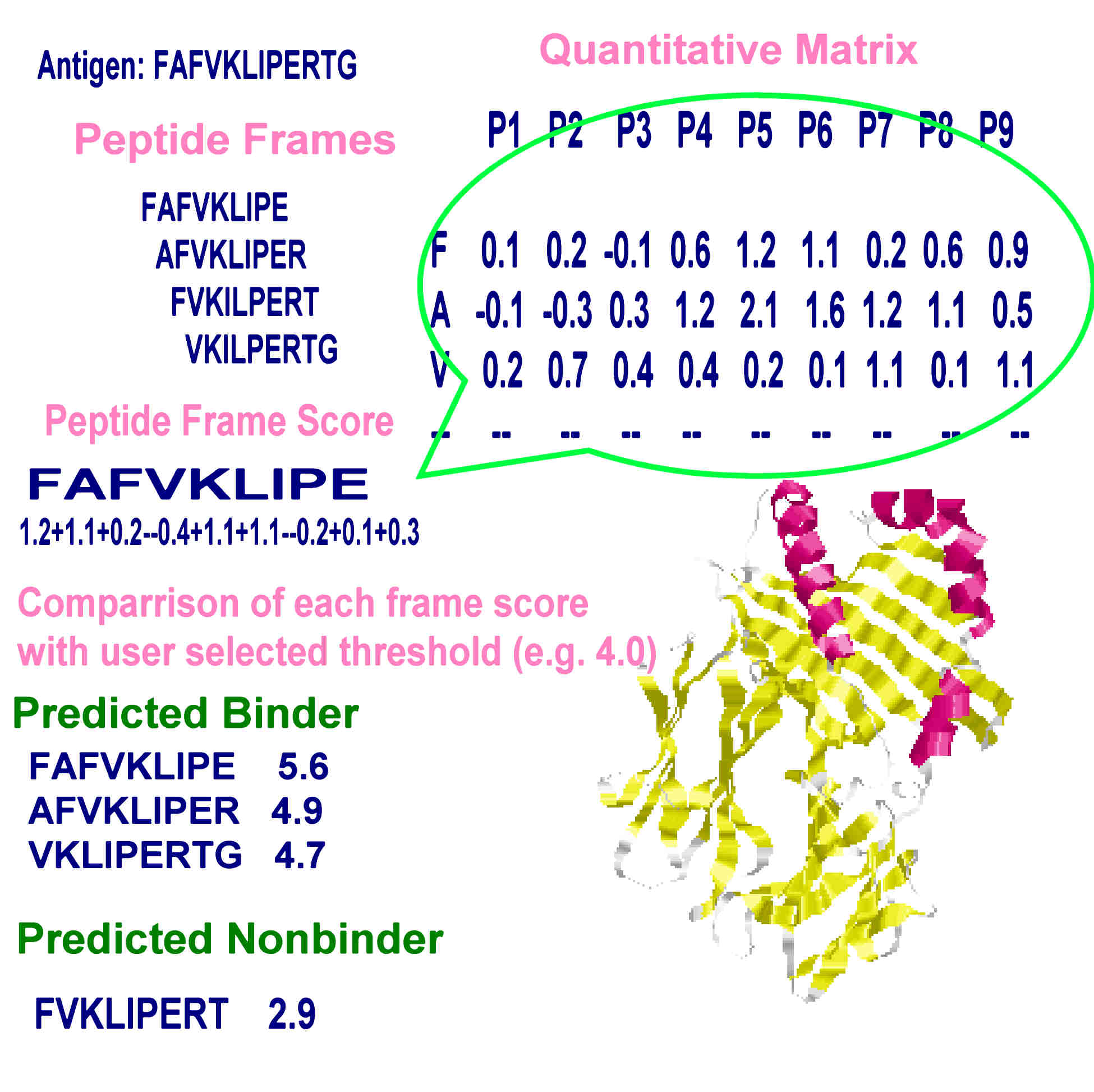
In silico design of MHC class I high binding affinity peptides through motifs activation map | BMC Bioinformatics | Full Text

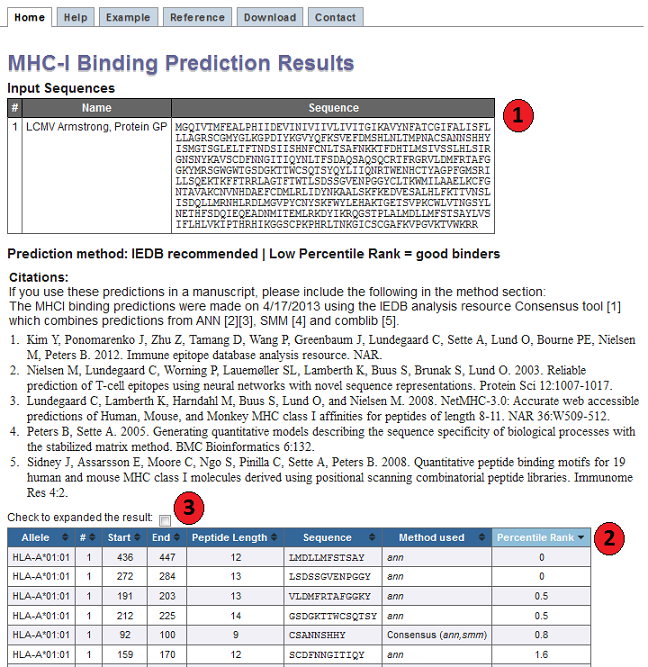
Flexible machine learning prediction of antigen presentation for rare and common HLA-I alleles | bioRxiv
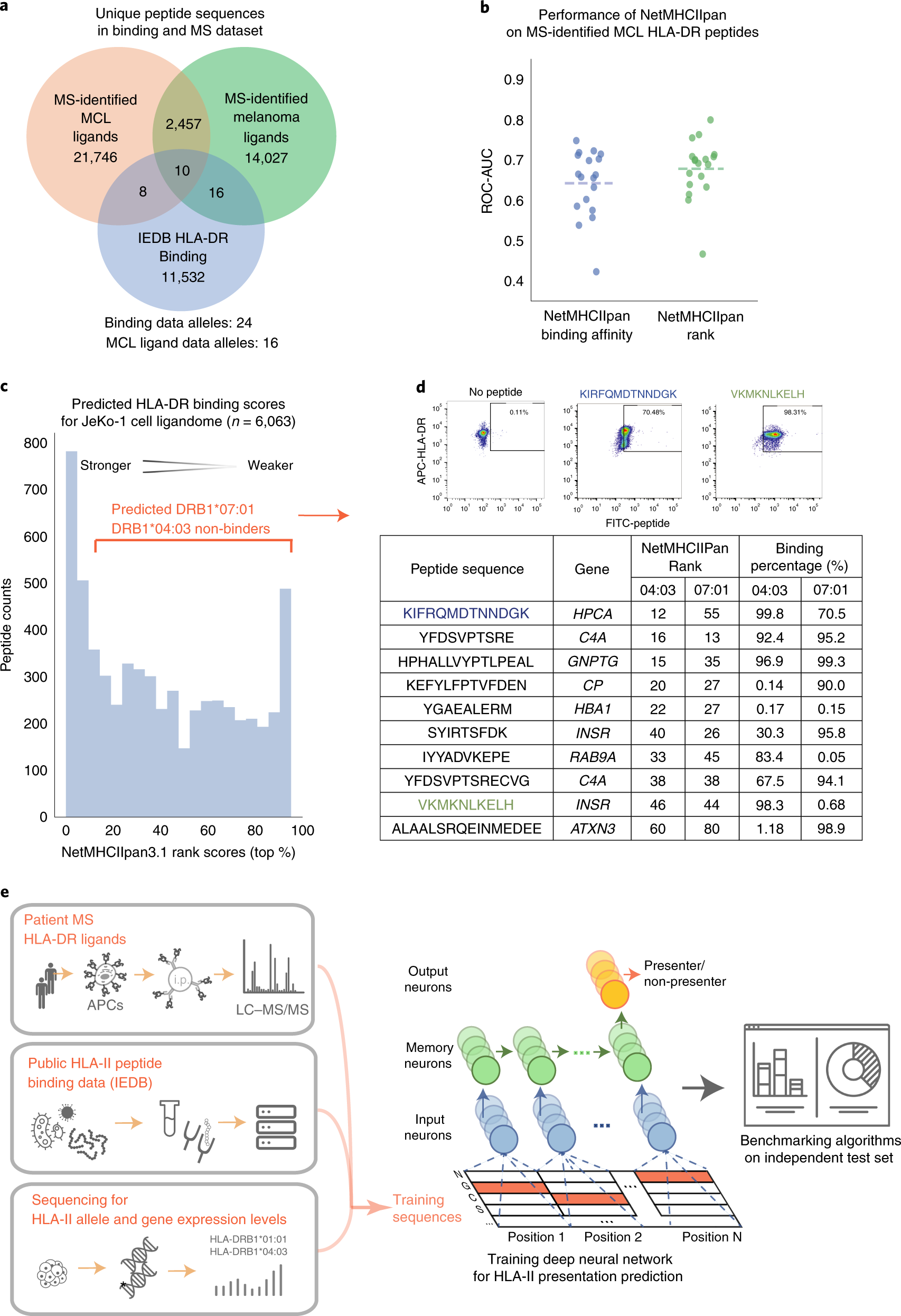
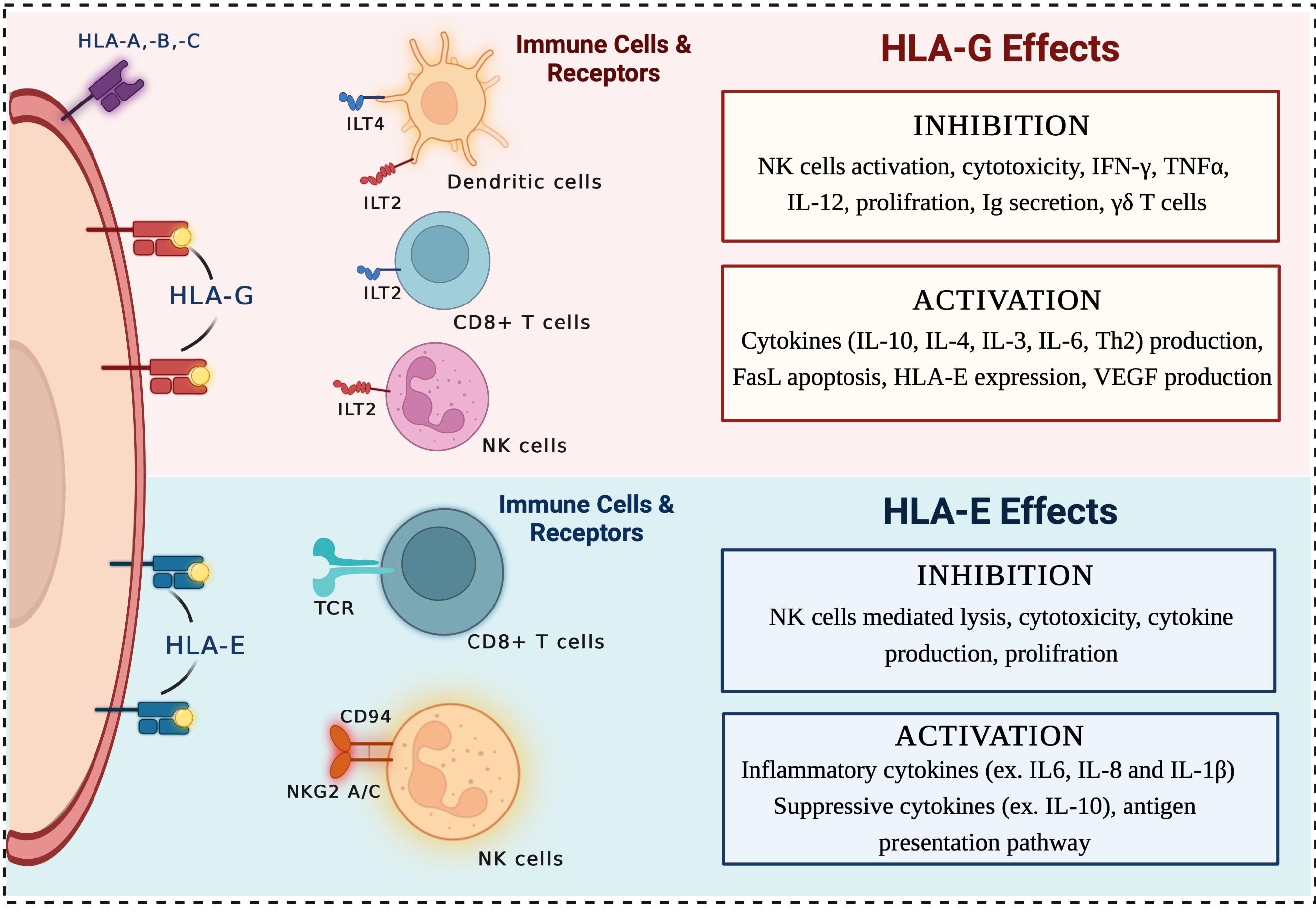
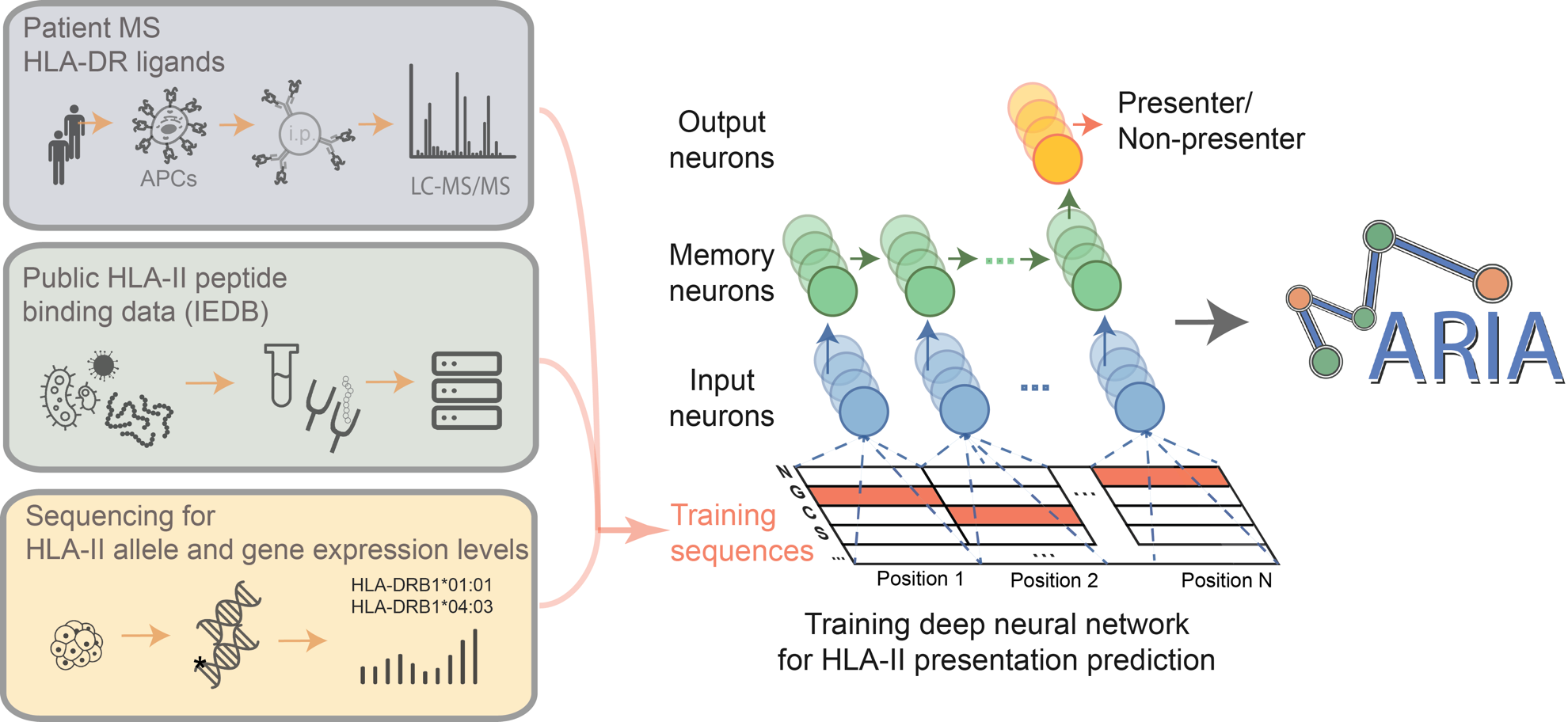
Predicting HLA class II antigen presentation through integrated deep learning | Nature Biotechnology
MHC Class II Binding Assays - ProImmune - Mastering Immunity _ MHC pentamers, CD1d tetramers, custom peptide synthesis, immunoassays, T cell epitope discovery, HLA tissue typing, cellular analysis, ligand binding assays, immune response

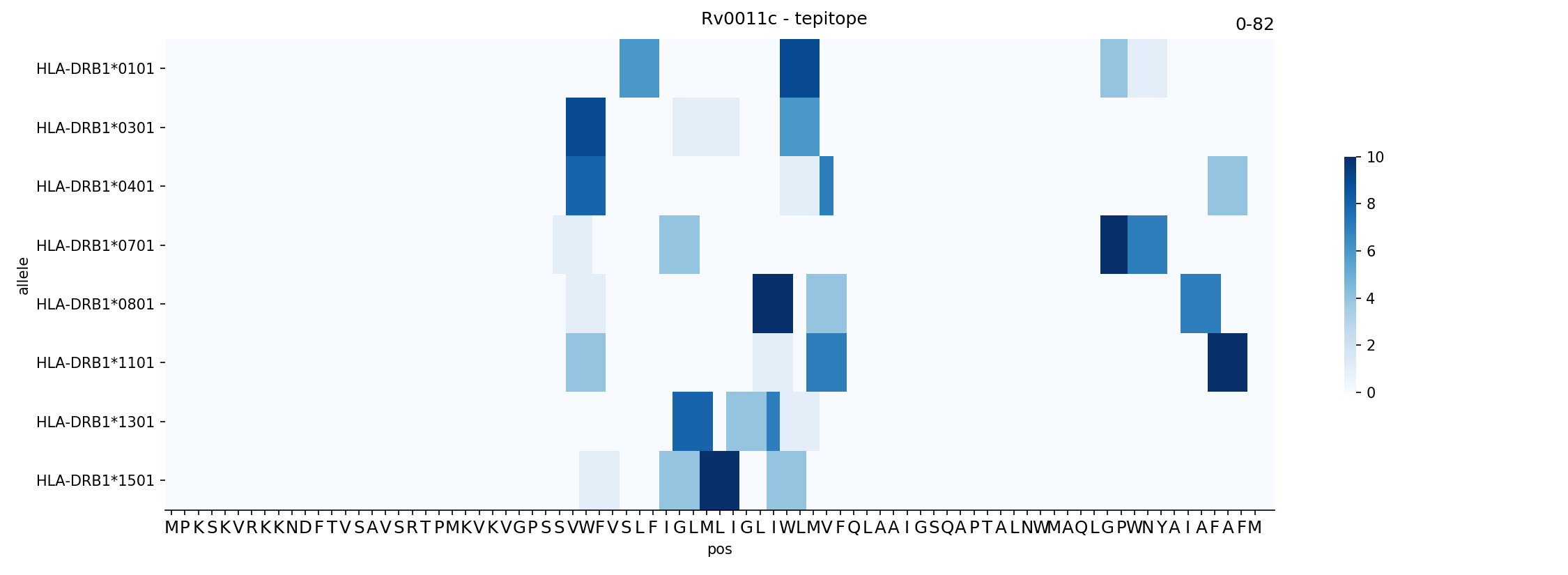
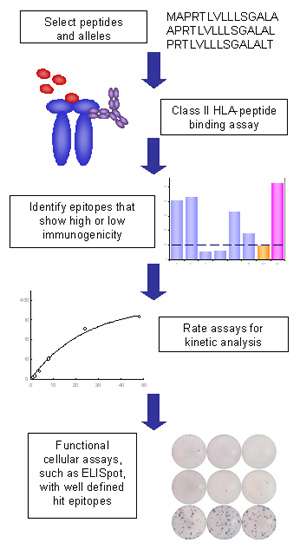
Prediction of HLA class II-restricted T cell epitopes. A strategy to... | Download Scientific Diagram
Quantitative Predictions of Peptide Binding to Any HLA-DR Molecule of Known Sequence: NetMHCIIpan | PLOS Computational Biology

Improved methods for predicting peptide binding affinity to MHC class II molecules - Jensen - 2018 - Immunology - Wiley Online Library

Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes - ScienceDirect

Combined assessment of MHC binding and antigen abundance improves T cell epitope predictions - ScienceDirect