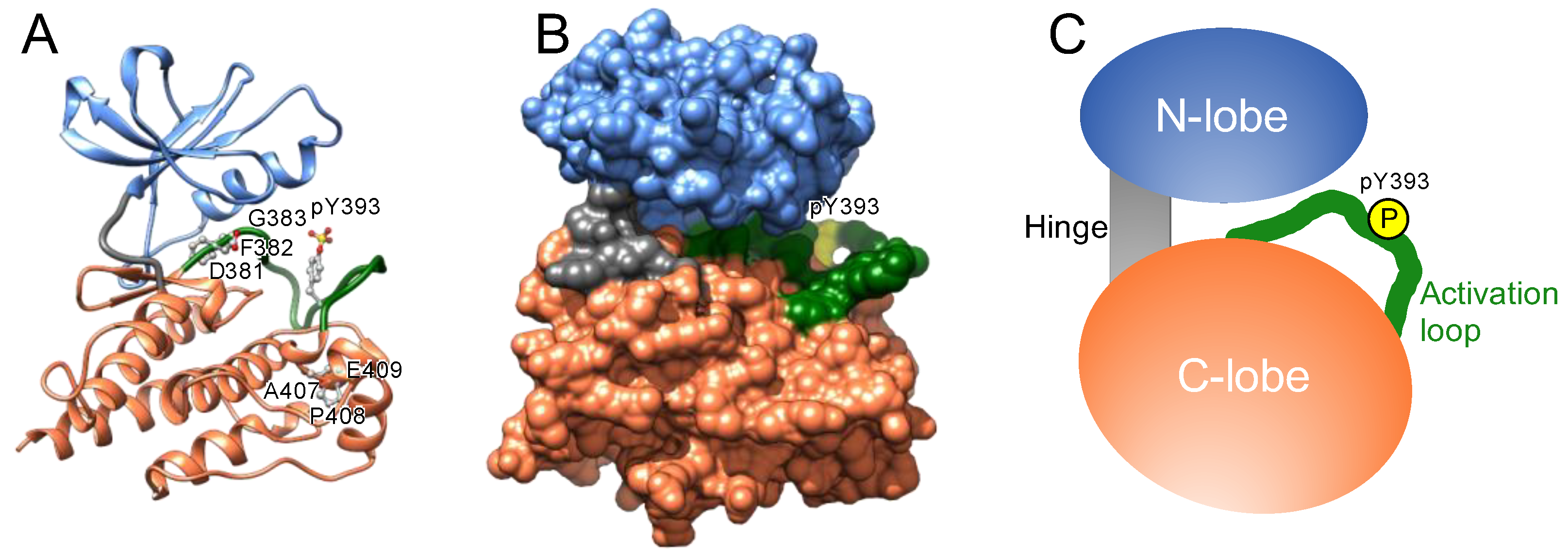
A) Structure of the kinase hinge binding motif with ATP illustrated by... | Download Scientific Diagram
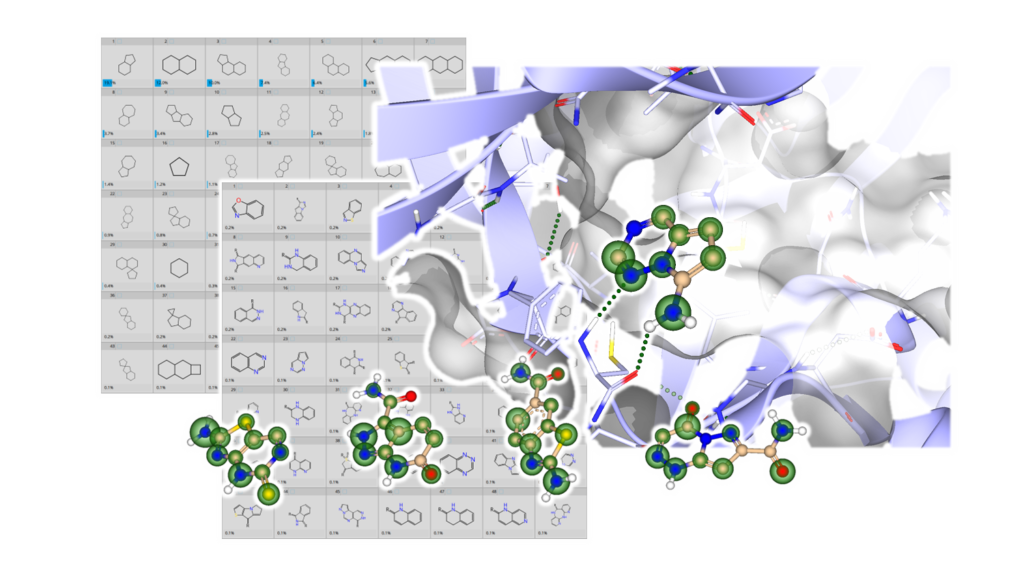
![PDF] De Novo Design of Protein Kinase Inhibitors by in Silico Identification of Hinge Region-Binding Fragments | Semantic Scholar PDF] De Novo Design of Protein Kinase Inhibitors by in Silico Identification of Hinge Region-Binding Fragments | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e1f213c145398224554d9b0983d883082be0cda7/5-Figure4-1.png)
PDF] De Novo Design of Protein Kinase Inhibitors by in Silico Identification of Hinge Region-Binding Fragments | Semantic Scholar

Figure 1 from De Novo Design of Protein Kinase Inhibitors by in Silico Identification of Hinge Region-Binding Fragments | Semantic Scholar

Kinase Crystal Miner: A Powerful Approach to Repurposing 3D Hinge Binding Fragments and Its Application to Finding Novel Bruton Tyrosine Kinase Inhibitors | Journal of Chemical Information and Modeling

Efficient Hit-to-Lead Searching of Kinase Inhibitor Chemical Space via Computational Fragment Merging | bioRxiv

Fragment Binding to Kinase Hinge: If Charge Distribution and Local pKa Shifts Mislead Popular Bioisosterism Concepts - Oebbeke - 2021 - Angewandte Chemie International Edition - Wiley Online Library

Kinase Crystal Miner: A Powerful Approach to Repurposing 3D Hinge Binding Fragments and Its Application to Finding Novel Bruton Tyrosine Kinase Inhibitors | Journal of Chemical Information and Modeling

Efficient Hit-to-Lead Searching of Kinase Inhibitor Chemical Space via Computational Fragment Merging | bioRxiv

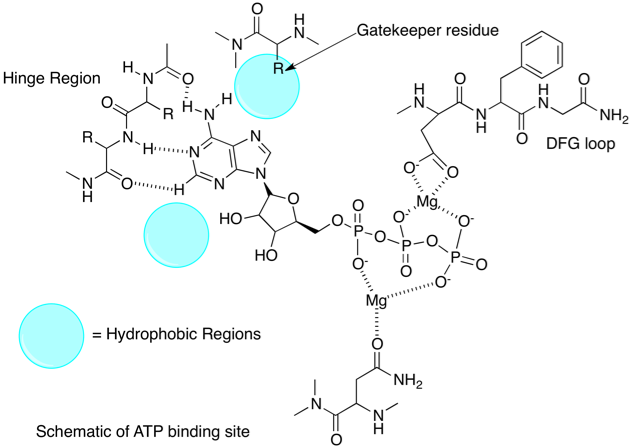
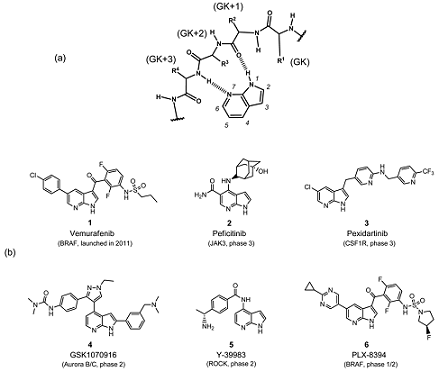
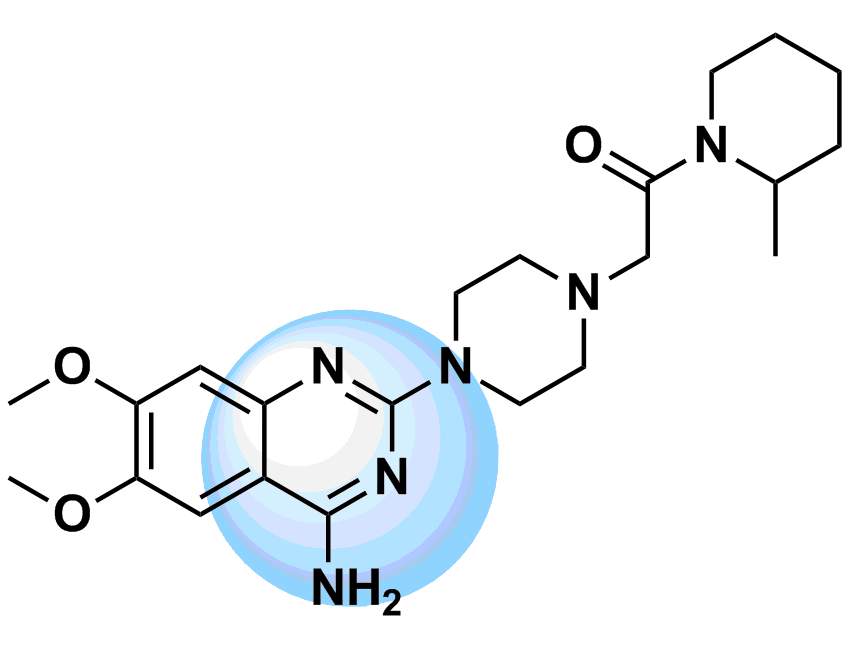
Designing of kinase hinge binders: A medicinal chemistry perspective - Sharma - 2022 - Chemical Biology & Drug Design - Wiley Online Library
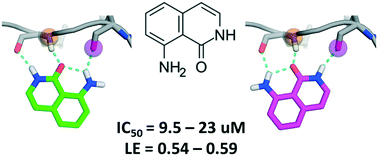
Discovery of a novel kinase hinge binder fragment by dynamic undocking - RSC Medicinal Chemistry (RSC Publishing)
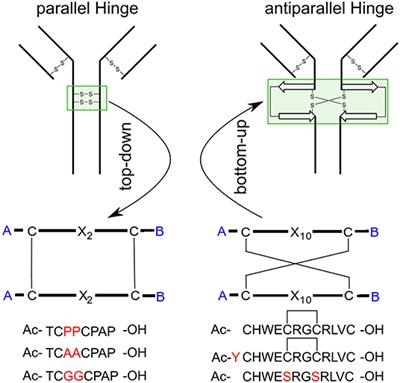
Frontiers | Comparing the Hinge-Type Mobility of Natural and Designed Intermolecular Bi-disulfide Domains

Molecular basis for the disruption of Keap1–Nrf2 interaction via Hinge & Latch mechanism | Communications Biology