Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase | Nature Communications

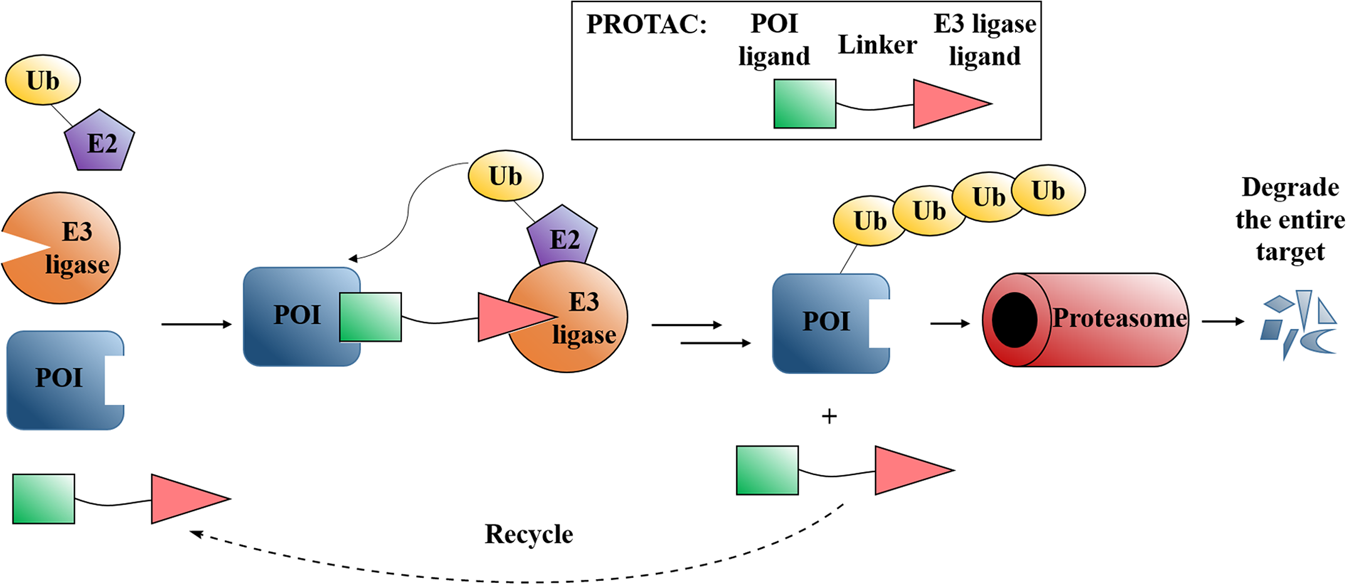
Computer aided drug design in the development of proteolysis targeting chimeras - Computational and Structural Biotechnology Journal

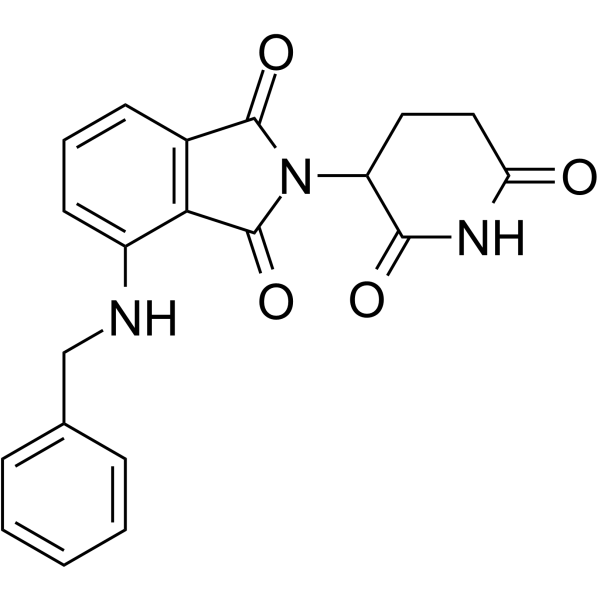
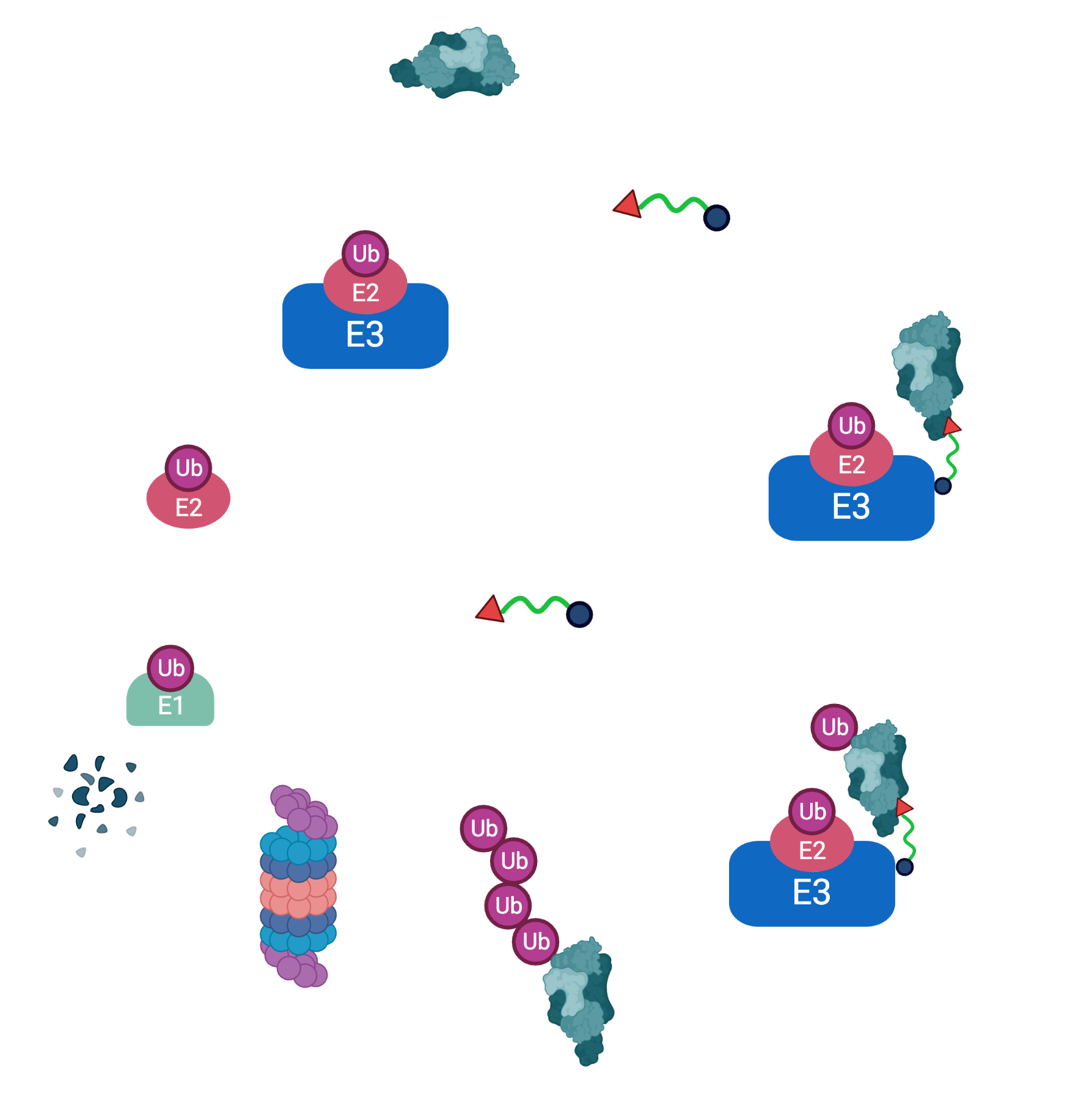
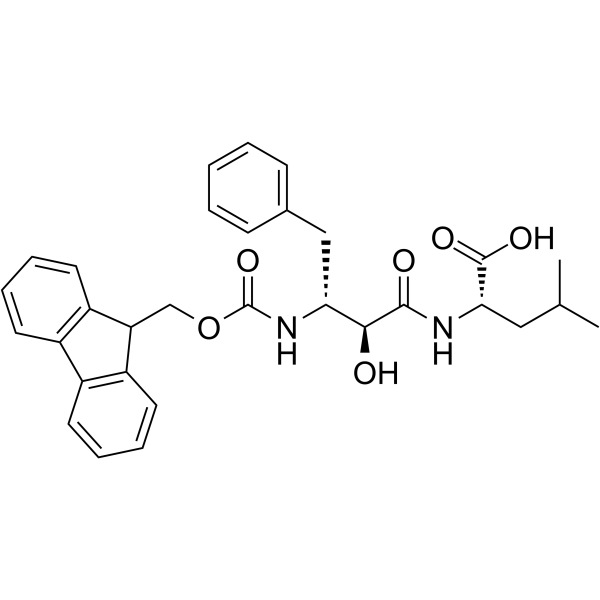
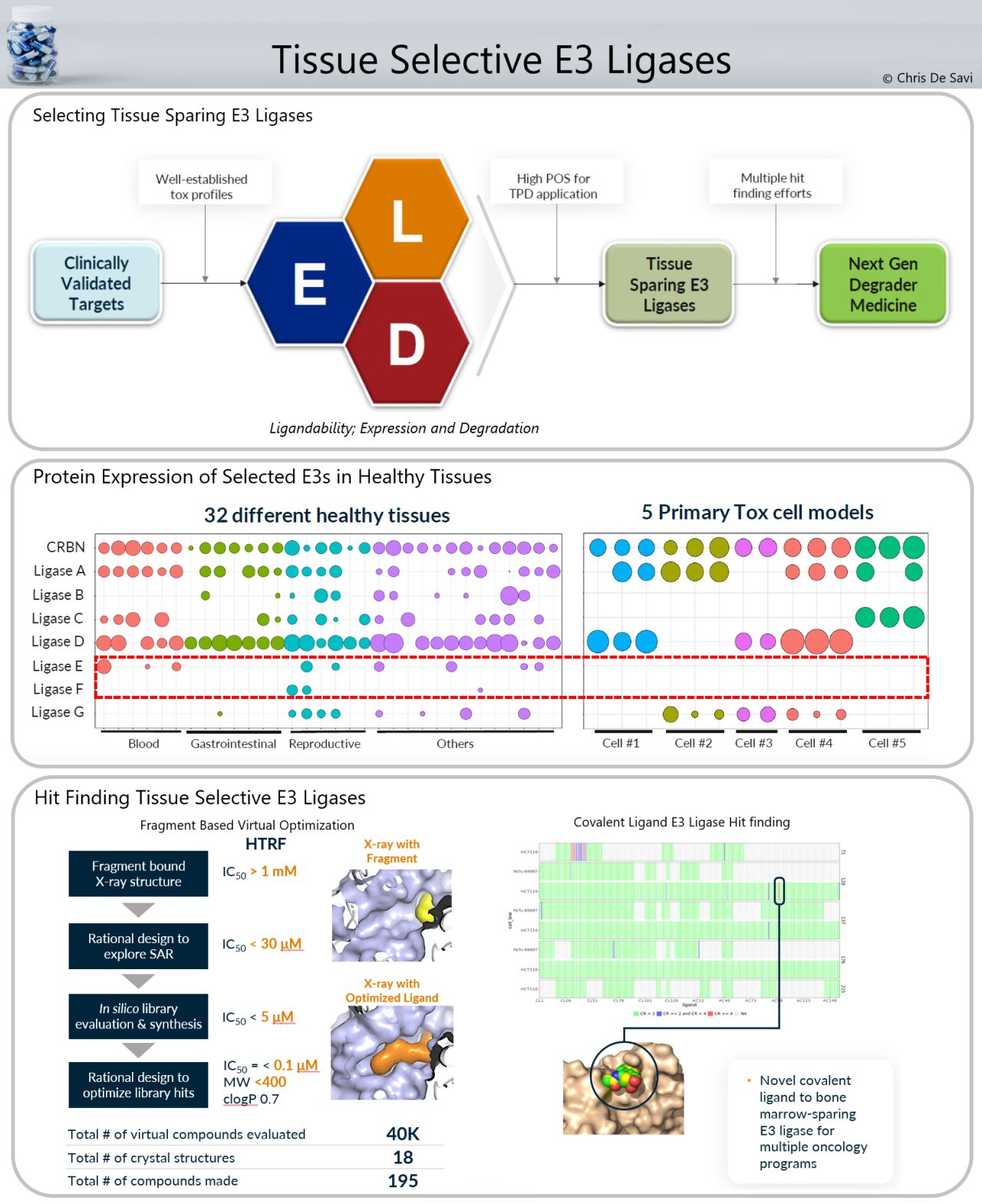
E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones - Tasuku Ishida, Alessio Ciulli, 2021
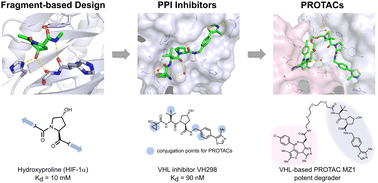
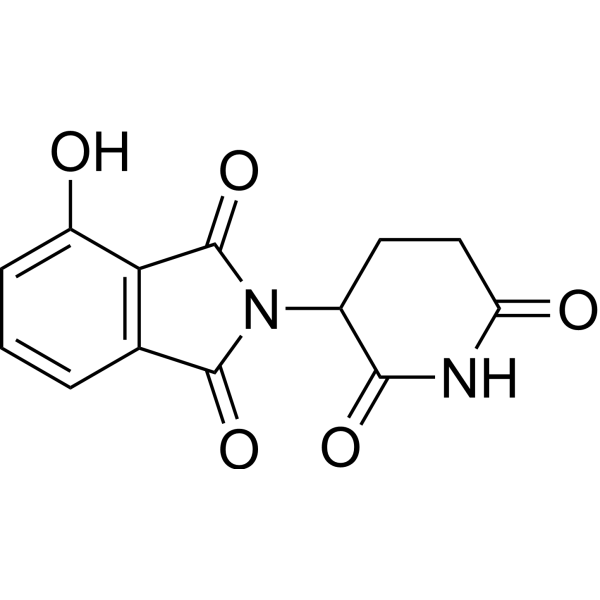
Discovery of small molecule ligands for the von Hippel-Lindau (VHL) E3 ligase and their use as inhibitors and PROTAC degraders - Chemical Society Reviews (RSC Publishing)
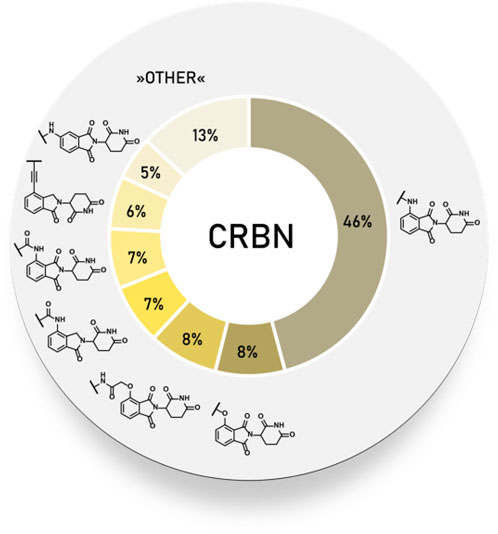
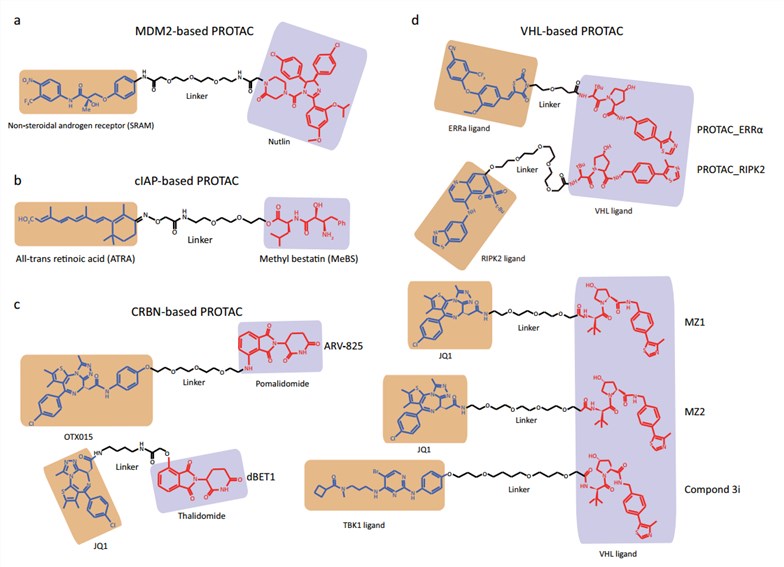
Frontiers | E3 Ligase Ligands in Successful PROTACs: An Overview of Syntheses and Linker Attachment Points
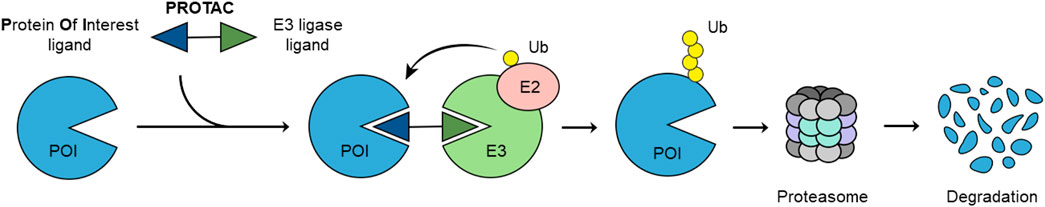
Frontiers | E3 Ligase Ligands in Successful PROTACs: An Overview of Syntheses and Linker Attachment Points

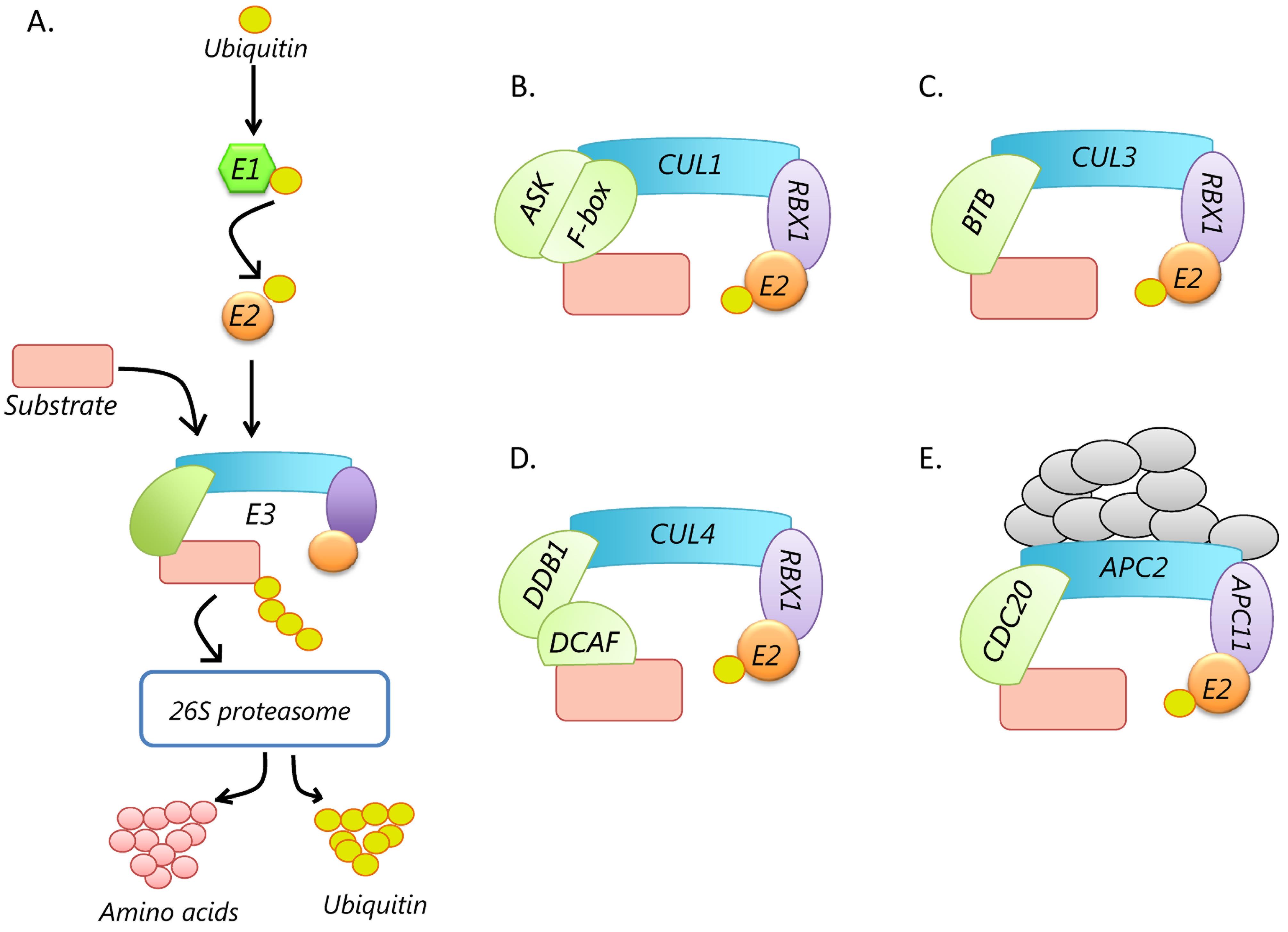
Beute für das Proteasom: Gezielter Proteinabbau aus medizinalchemischer Perspektive - Luh - 2020 - Angewandte Chemie - Wiley Online Library

Electrophilic Screening Platforms for Identifying Novel Covalent Ligands for E3 Ligases | Biochemistry
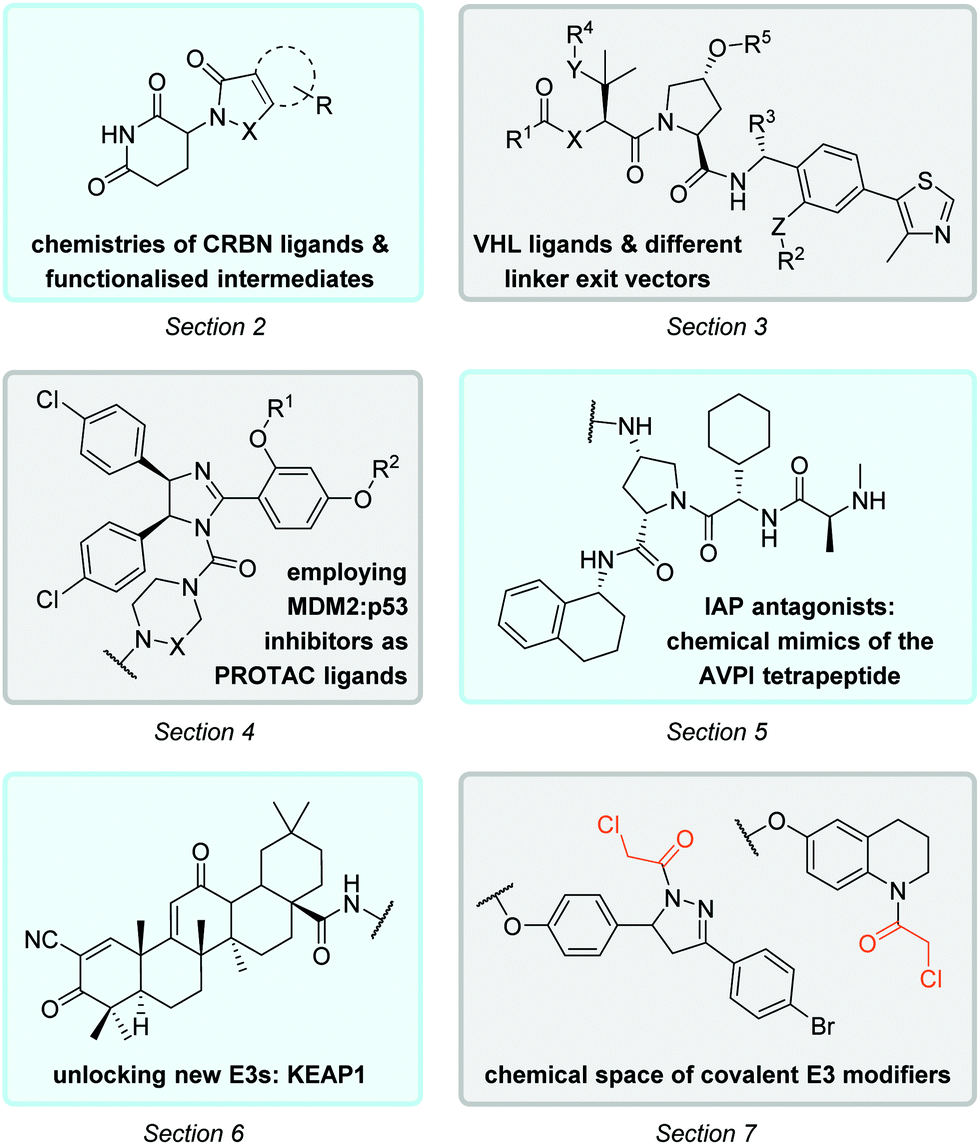
E3 ligase ligand chemistries: from building blocks to protein degraders - Chemical Society Reviews (RSC Publishing) DOI:10.1039/D2CS00148A