Predicting Hot Spot Residues at Protein–DNA Binding Interfaces Based on Sequence Information | Interdisciplinary Sciences: Computational Life Sciences
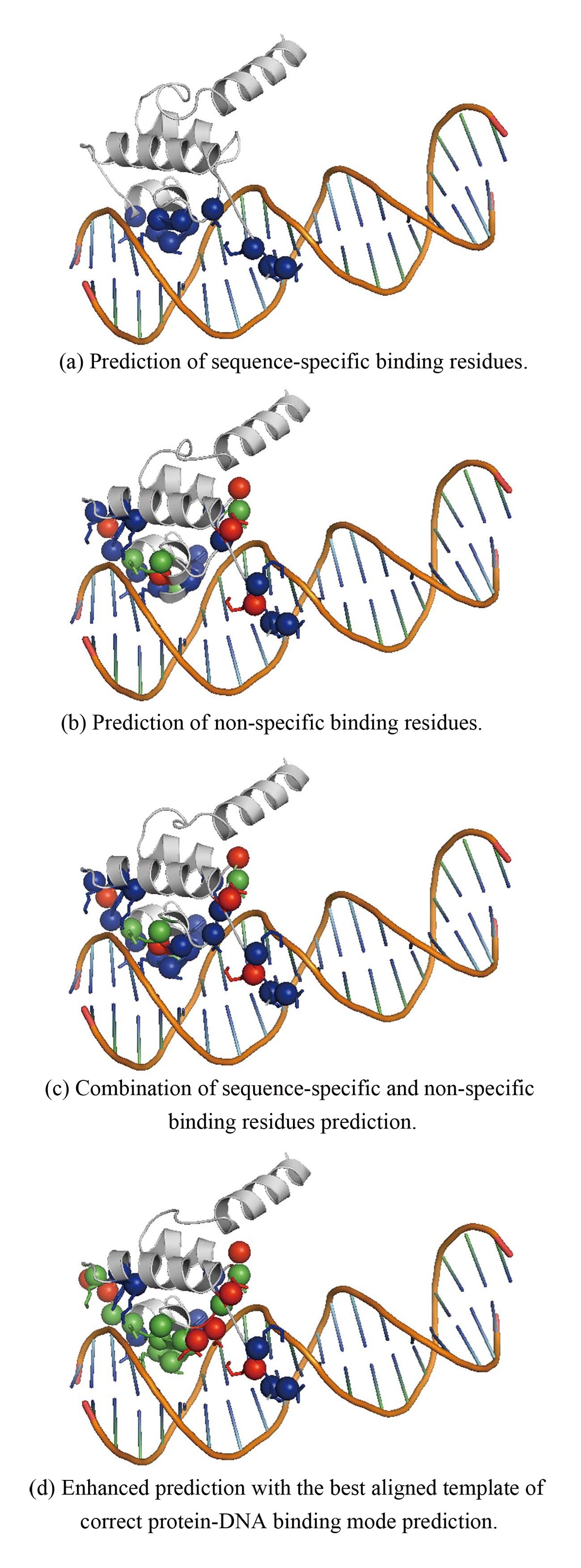
DNA-binding residues and binding mode prediction with binding-mechanism concerned models | BMC Genomics | Full Text

Examples of DNA-binding protein prediction on APO104. (A–D) In each... | Download Scientific Diagram
![PDF] DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. | Semantic Scholar PDF] DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/5d68bdf0391af25147a48171a54fea49d64d16d4/2-Figure1-1.png)
PDF] DNA Shape Features Improve Transcription Factor Binding Site Predictions In Vivo. | Semantic Scholar
GitHub - biomed-AI/GraphSite: GraphSite: protein-DNA binding site prediction using graph transformer and predicted protein structures

Improving DNA-Binding Protein Prediction Using Three-Part Sequence-Order Feature Extraction and a Deep Neural Network Algorithm | Journal of Chemical Information and Modeling

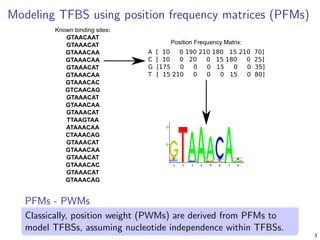
Modeling binding specificities of transcription factor pairs with random forests | BMC Bioinformatics | Full Text

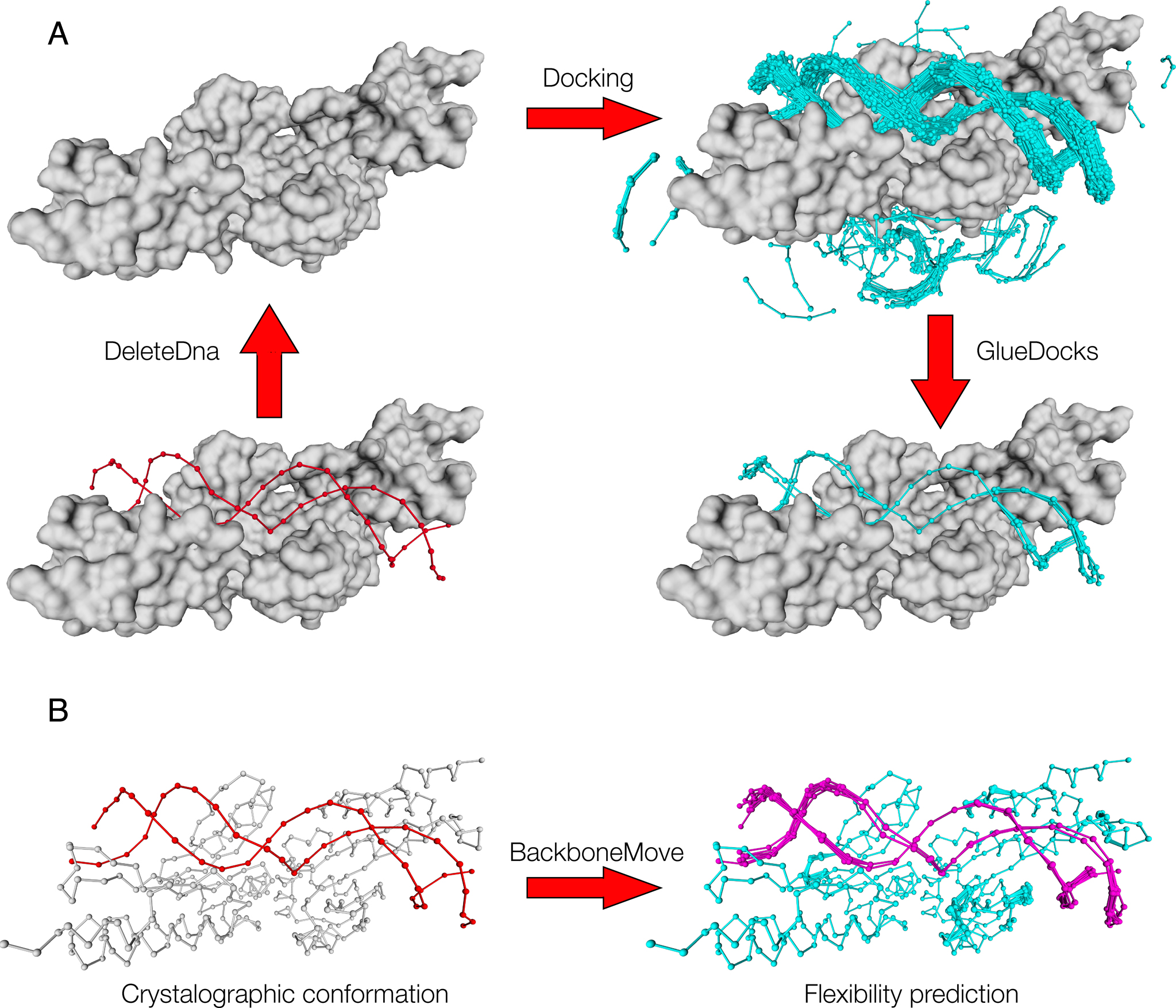
An efficient algorithm for improving structure-based prediction of transcription factor binding sites | BMC Bioinformatics | Full Text

DeepGRN: prediction of transcription factor binding site across cell-types using attention-based deep neural networks | BMC Bioinformatics | Full Text
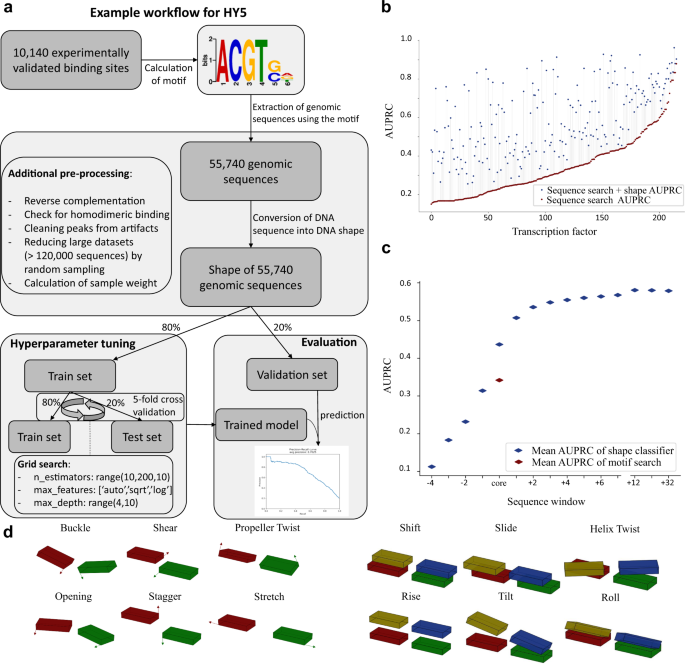
Local DNA shape is a general principle of transcription factor binding specificity in Arabidopsis thaliana | Nature Communications

Improving the prediction of DNA-protein binding by integrating multi-scale dense convolutional network with fault-tolerant coding - ScienceDirect

Four major steps in structure-based prediction of transcription factor... | Download Scientific Diagram
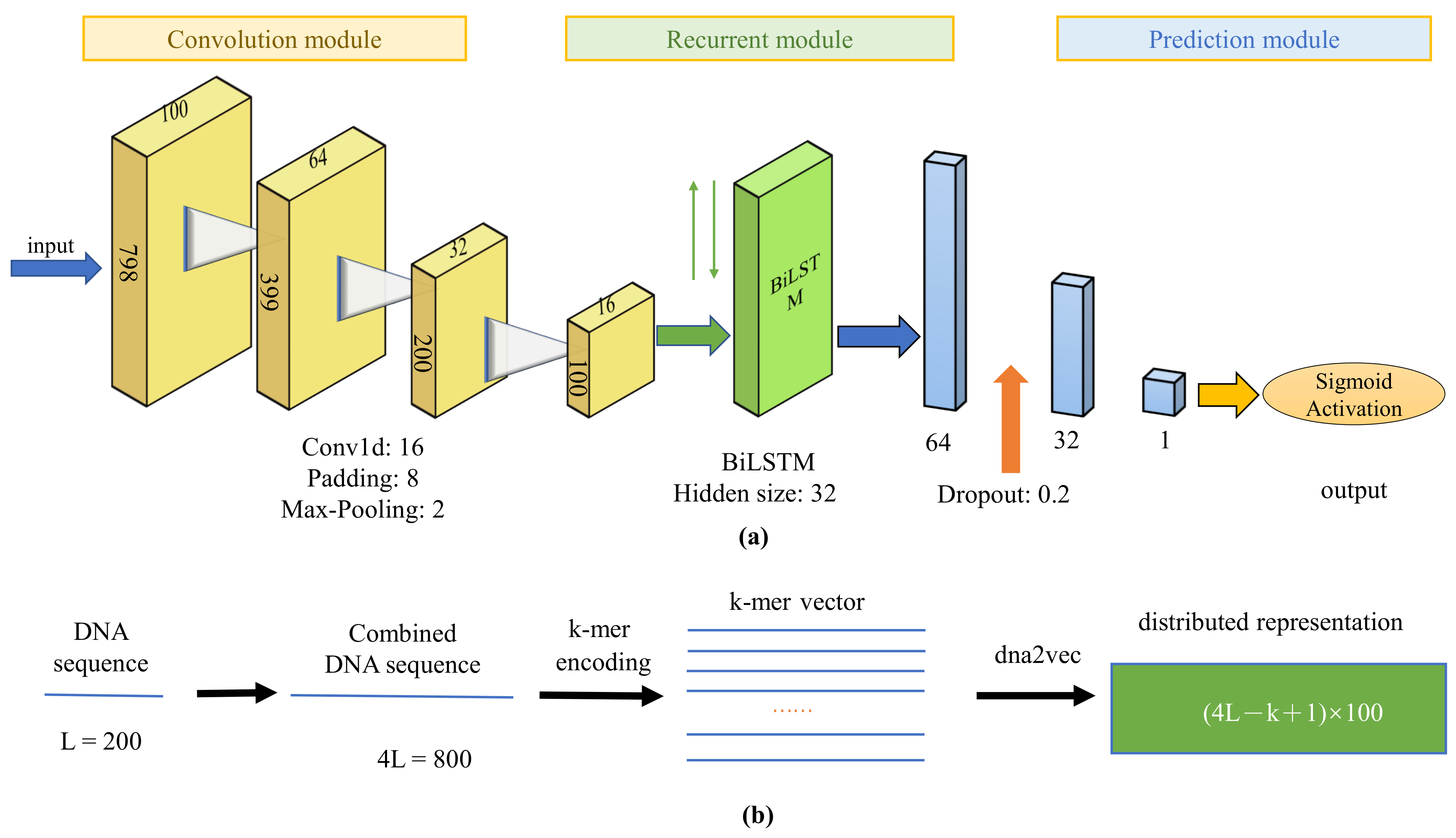
IJMS | Free Full-Text | DeepD2V: A Novel Deep Learning-Based Framework for Predicting Transcription Factor Binding Sites from Combined DNA Sequence
![Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ] Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2021/11262/1/fig-1-2x.jpg)
Prediction of DNA binding proteins using local features and long-term dependencies with primary sequences based on deep learning [PeerJ]