Lipid II-binding antibiotics – differential antibiotic activities beyond sequestration of the central peptidoglycan precursor
Analysis of differential binding across cell lines: ChIP-seq signals... | Download Scientific Diagram

Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation | Genome Biology | Full Text

Identification of ligand binding sites in intrinsically disordered proteins with a differential binding score | Scientific Reports
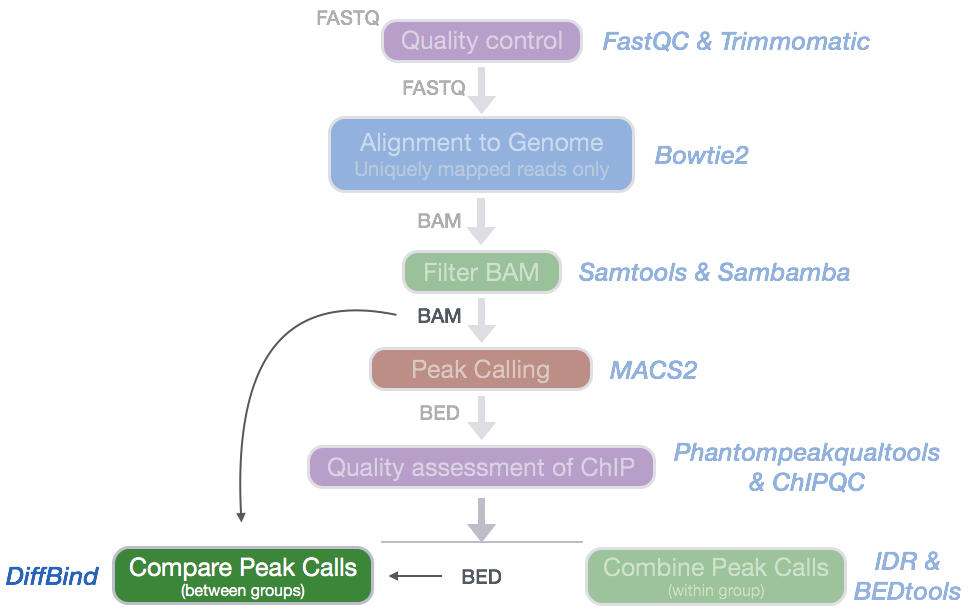
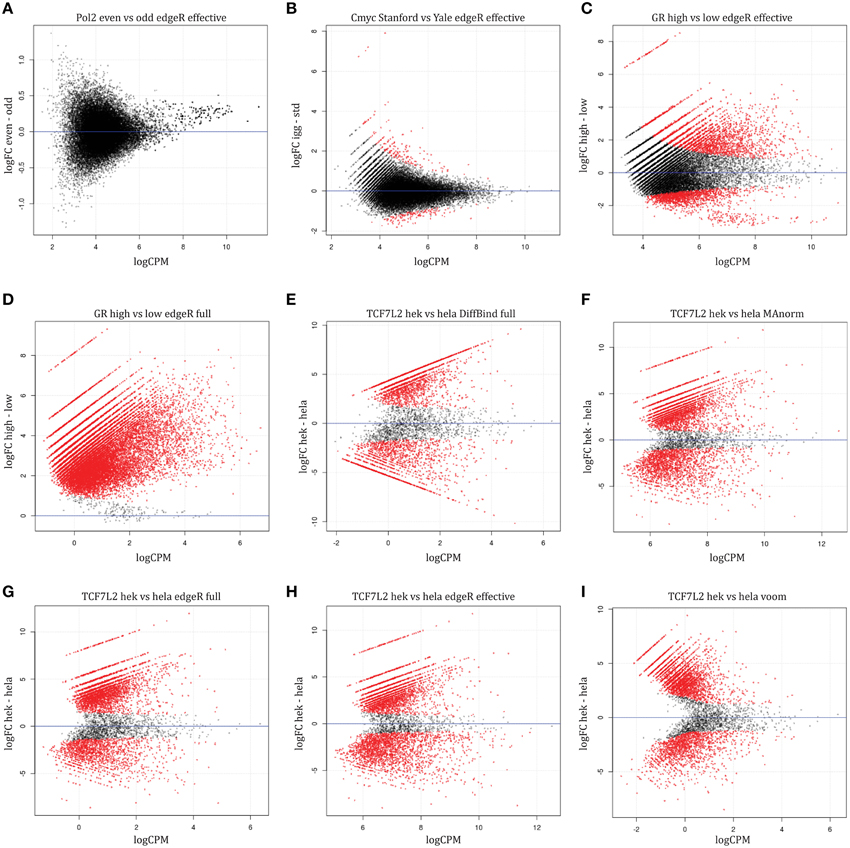
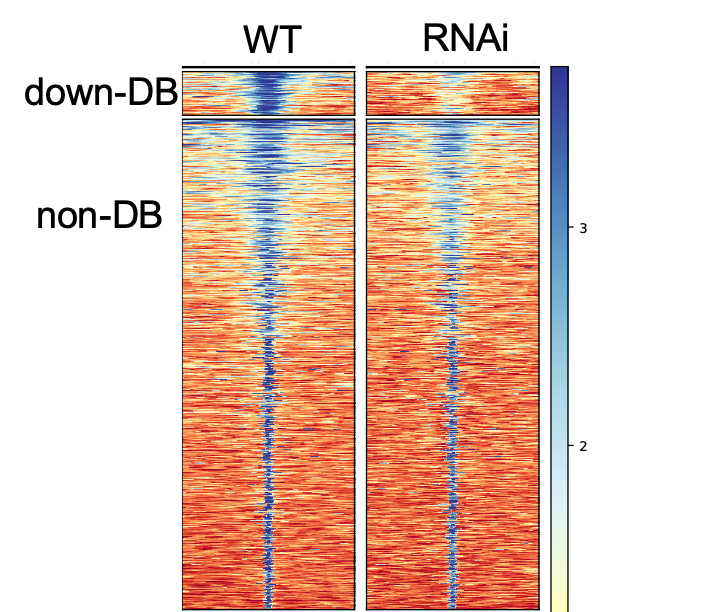
Differential Peak calling using DiffBind | Introduction to ChIP-Seq using high-performance computing

pH Dependent Differential Binding Behavior of Prtotease Inhibitor Molecular Drugs for SARS-COV-2 | Biological and Medicinal Chemistry | ChemRxiv | Cambridge Open Engage
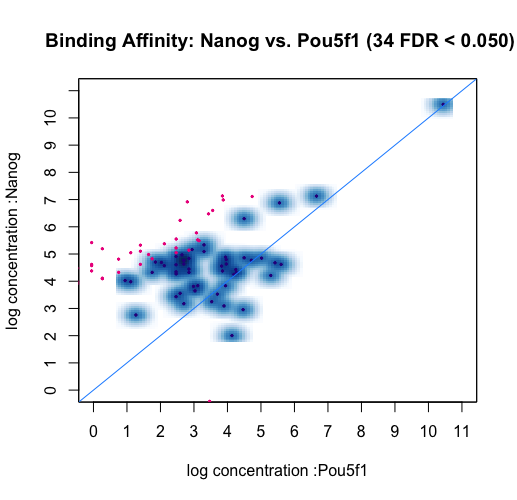
Differential Peak calling using DiffBind | Introduction to ChIP-Seq using high-performance computing

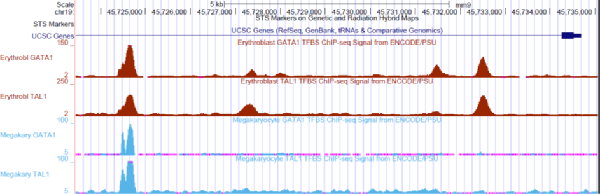
Divergence in DNA Specificity among Paralogous Transcription Factors Contributes to Their Differential In Vivo Binding - ScienceDirect

a. Decomposition of the differential binding energy into interaction... | Download Scientific Diagram

File:Exploring-the-Origin-of-Differential-Binding-Affinities-of-Human-Tubulin-Isotypes-αβII-αβIII-and-pone.0156048.s012.ogv - Wikimedia Commons