Benefiting from the intrinsic role of epigenetics to predict patterns of CTCF binding - Computational and Structural Biotechnology Journal

Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver. - Abstract - Europe PMC

CTCF: the protein, the binding partners, the binding sites and their chromatin loops | Philosophical Transactions of the Royal Society B: Biological Sciences

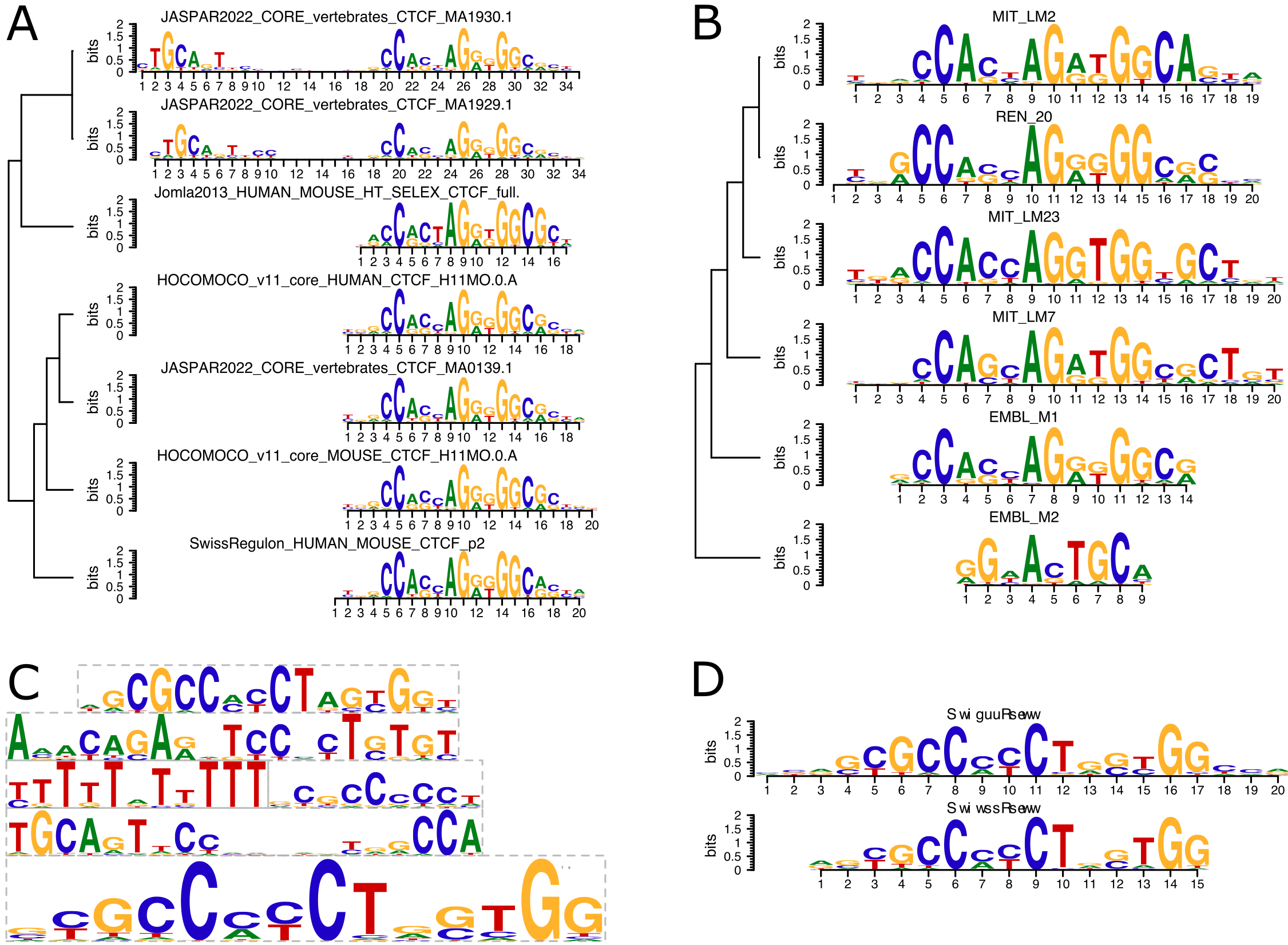
Sequence logo and mutual information. Figure a) depicts the sequence... | Download Scientific Diagram

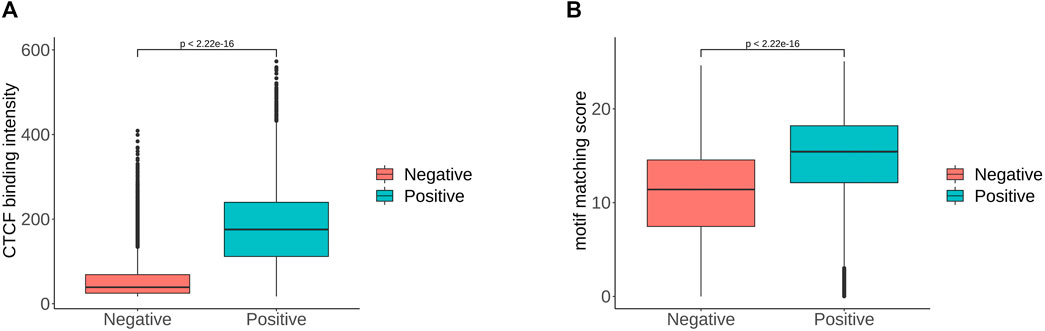
Predicting CTCF-mediated chromatin interactions by integrating genomic and epigenomic features | Nature Communications

Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains | Nature Communications

Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation | Genome Biology | Full Text

Detection and conservation of CTCF-binding sites. (a) Number of CTCF... | Download Scientific Diagram

Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation | Genome Biology | Full Text

Motif logos for CTCF sites derived separately from the best scoring... | Download Scientific Diagram
CTCF Genomic Binding Sites in Drosophila and the Organisation of the Bithorax Complex | PLOS Genetics

CTCF: the protein, the binding partners, the binding sites and their chromatin loops | Philosophical Transactions of the Royal Society B: Biological Sciences

Low-affinity CTCF binding drives transcriptional regulation whereas high-affinity binding encompasses architectural functions - ScienceDirect