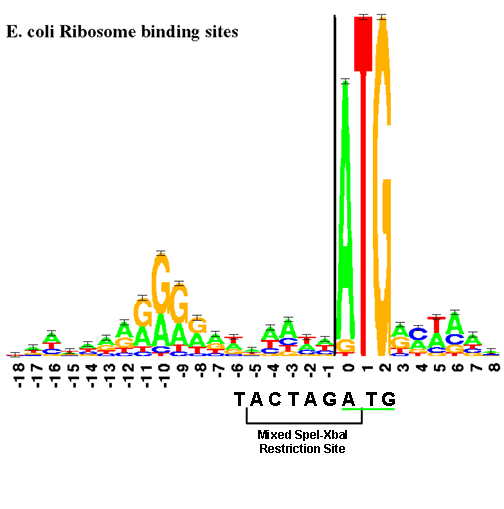
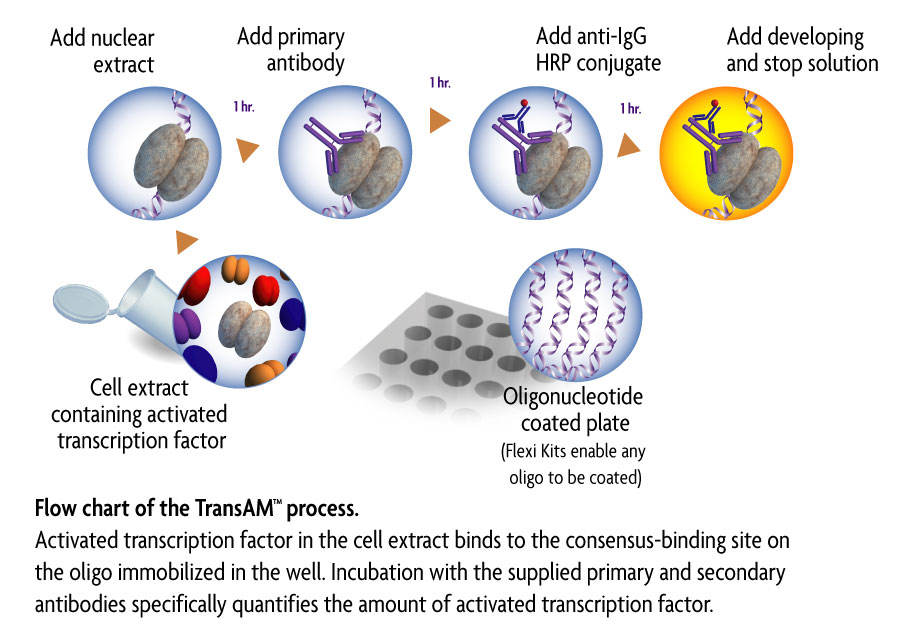
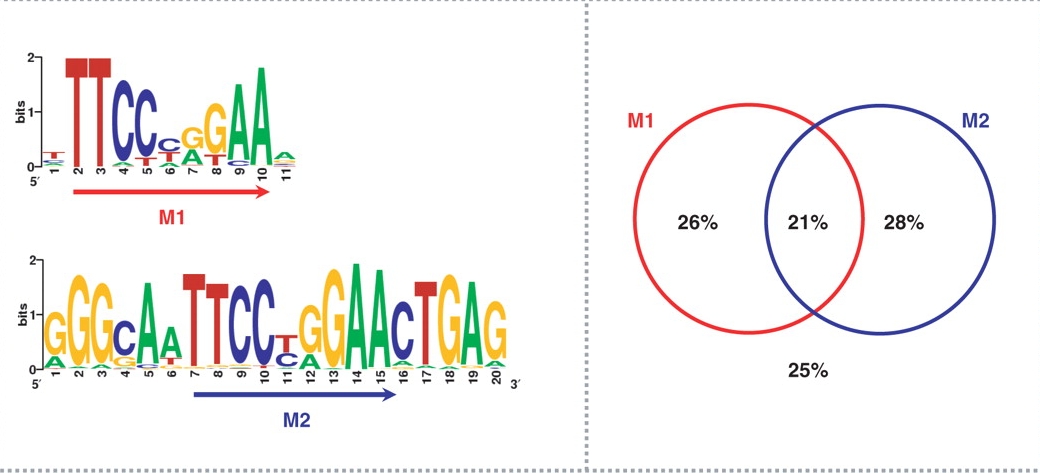
Flanking sequence context-dependent transcription factor binding in early Drosophila development | BMC Bioinformatics | Full Text

FleQ DNA Binding Consensus Sequence Revealed by Studies of FleQ-Dependent Regulation of Biofilm Gene Expression in Pseudomonas aeruginosa | Journal of Bacteriology

An in vitro technique to identify the RNA binding-site sequences for RNA- binding proteins | BioTechniques

DNA Consensus Sequence Motif for Binding Response Regulator PhoP, a Virulence Regulator of Mycobacterium tuberculosis | Biochemistry
Genome-wide global identification of NRF2 binding sites in A549 non-small cell lung cancer cells by ChIP-Seq reveals NRF2 regulation of genes involved in focal adhesion pathways | Aging

Genome-wide discovery of functional transcription factor binding sites by comparative genomics: The case of Stat3 | PNAS

Accuracy of inferred binding sites. Consensus binding motifs for CTCF,... | Download Scientific Diagram

DNA-binding properties of the MADS-domain transcription factor SEPALLATA3 and mutant variants characterized by SELEX-seq | Plant Molecular Biology

p63 consensus DNA-binding site: identification, analysis and application into a p63MH algorithm | Oncogene

MrpC binding site consensus sequences and the consensus sequence for... | Download Scientific Diagram

Systematic analysis of low-affinity transcription factor binding site clusters in vitro and in vivo establishes their functional relevance | bioRxiv

Consensus logo for human CREB binding sites (E < 3.8 x 10-125). This... | Download Scientific Diagram