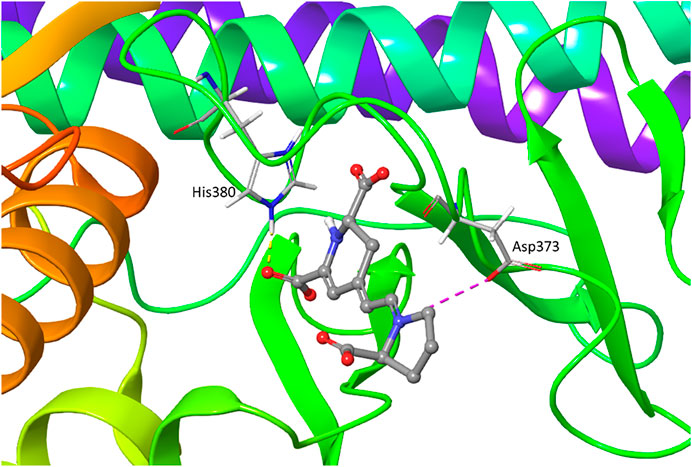
Binding pose metadynamics to identify the most stable orientation. (A)... | Download Scientific Diagram

Prediction of Protein–Ligand Binding Poses via a Combination of Induced Fit Docking and Metadynamics Simulations | Journal of Chemical Theory and Computation
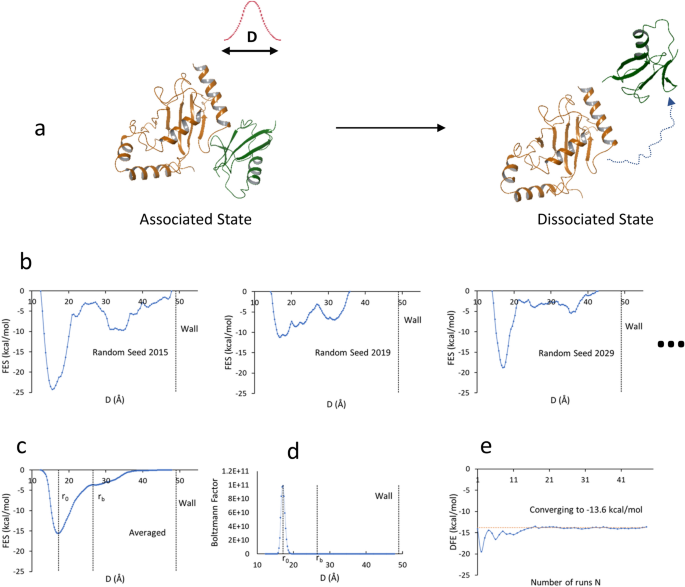
A highly accurate metadynamics-based Dissociation Free Energy method to calculate protein–protein and protein–ligand binding potencies | Scientific Reports
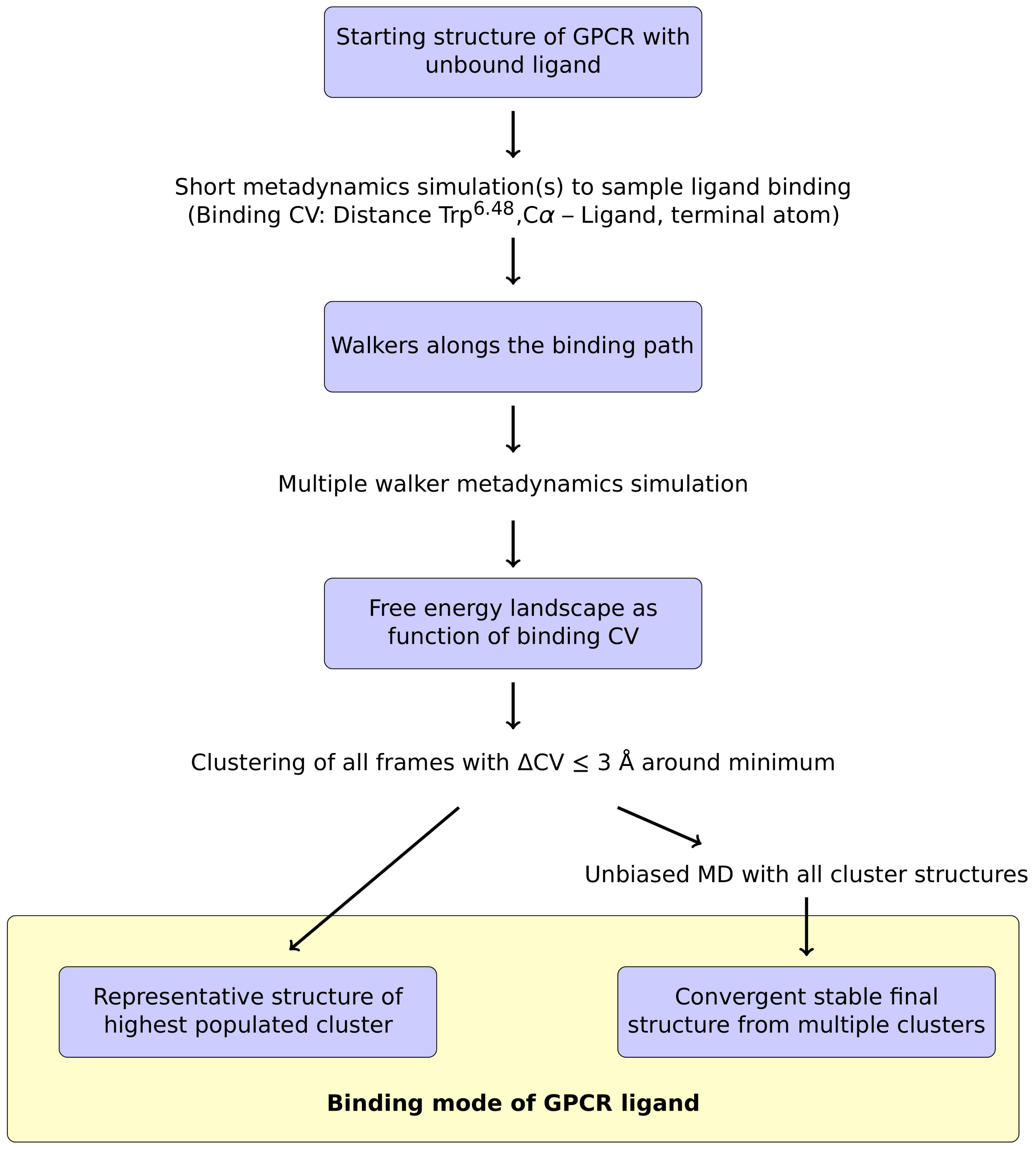
IJMS | Free Full-Text | A Metadynamics-Based Protocol for the Determination of GPCR-Ligand Binding Modes

Prediction of Protein–Ligand Binding Poses via a Combination of Induced Fit Docking and Metadynamics Simulations | Journal of Chemical Theory and Computation
![PDF) Binding pose analysis of hydroxyethylamine based β-secretase inhibitors and application thereof to the design and synthesis of novel indeno[1,2-b]indole based inhibitors PDF) Binding pose analysis of hydroxyethylamine based β-secretase inhibitors and application thereof to the design and synthesis of novel indeno[1,2-b]indole based inhibitors](https://i1.rgstatic.net/publication/346952643_Binding_pose_analysis_of_hydroxyethylamine_based_b-secretase_inhibitors_and_application_thereof_to_the_design_and_synthesis_of_novel_indeno12-bindole_based_inhibitors/links/5fe9180c92851c13fec99ed5/largepreview.png)
PDF) Binding pose analysis of hydroxyethylamine based β-secretase inhibitors and application thereof to the design and synthesis of novel indeno[1,2-b]indole based inhibitors

Prediction of Protein–Ligand Binding Poses via a Combination of Induced Fit Docking and Metadynamics Simulations | Journal of Chemical Theory and Computation

Binding pose metadynamics to identify the most stable orientation. (A)... | Download Scientific Diagram

Prediction of Protein–Ligand Binding Poses via a Combination of Induced Fit Docking and Metadynamics Simulations | Journal of Chemical Theory and Computation

Binding pose metadynamics results for complexes with ampicillin (left)... | Download Scientific Diagram

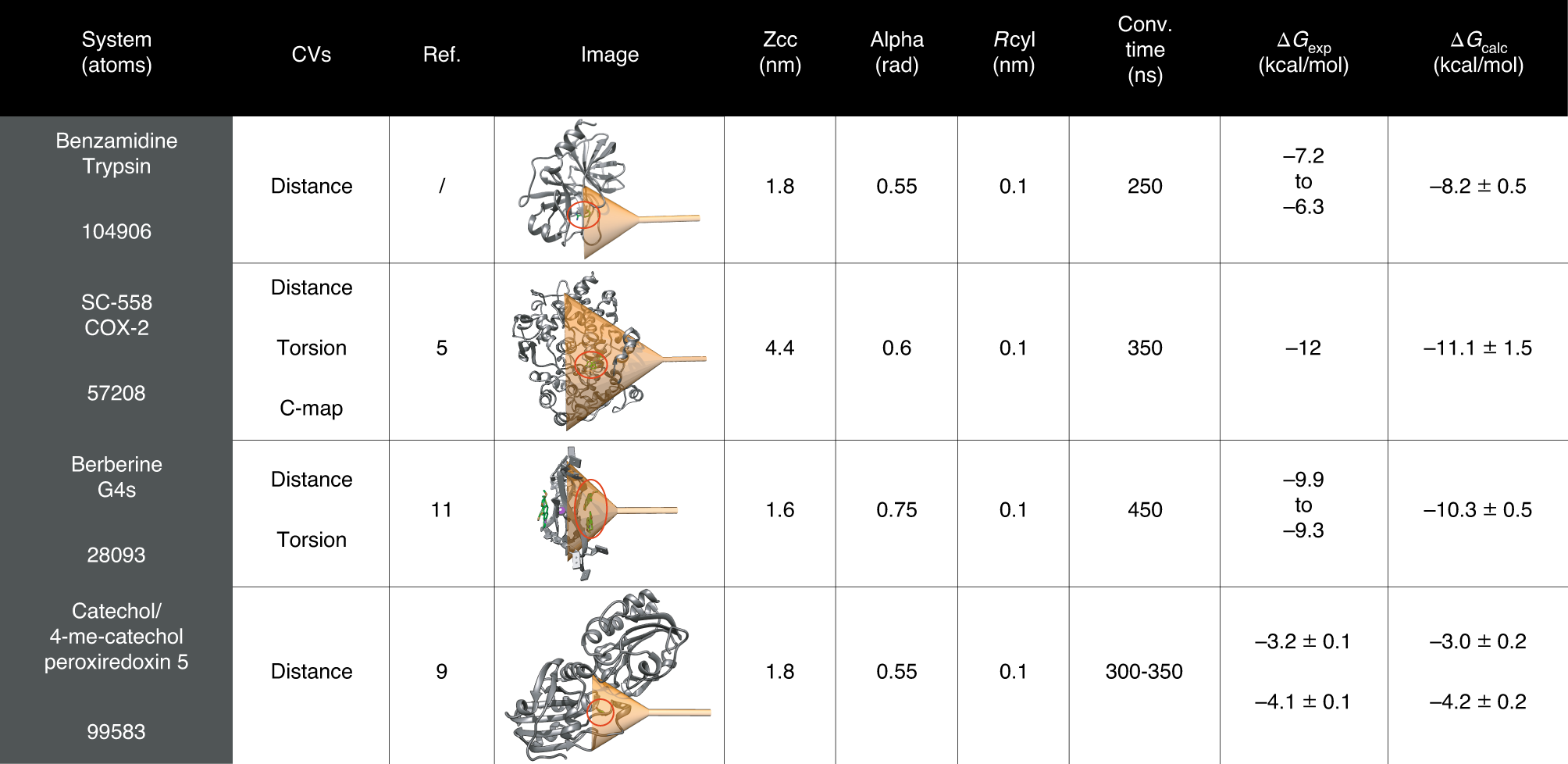
Metadynamics simulations of ligands binding to protein surfaces: a novel tool for rational drug design - Physical Chemistry Chemical Physics (RSC Publishing)

Alchemical free energy calculations via metadynamics: Application to the theophylline‐RNA aptamer complex - Tanida - 2020 - Journal of Computational Chemistry - Wiley Online Library
Prediction of Protein−Ligand Binding Poses via a Combination of Induced Fit Docking and Metadynamics Simulations
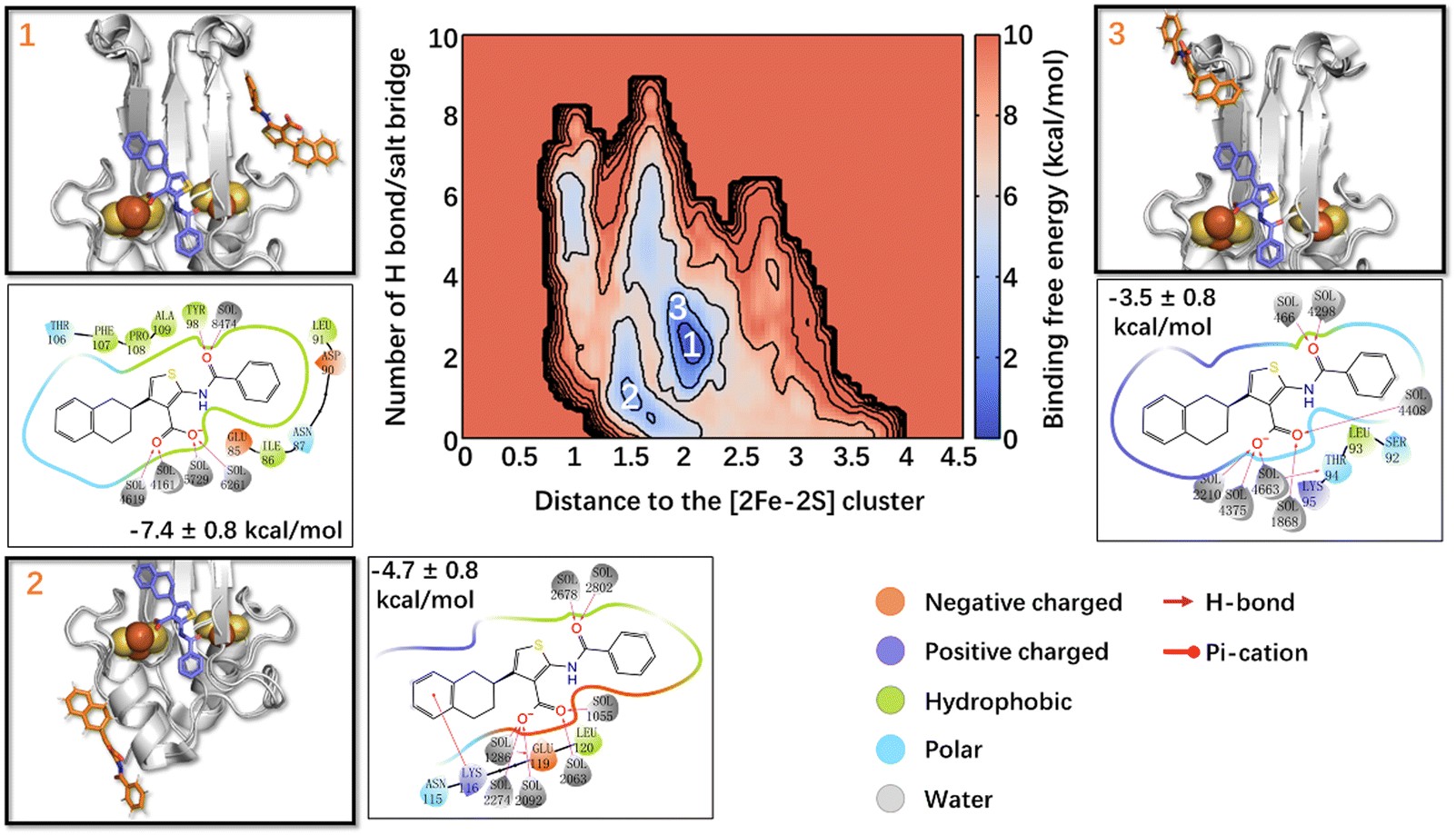
Metadynamics simulations of ligands binding to protein surfaces: a novel tool for rational drug design - Physical Chemistry Chemical Physics (RSC Publishing) DOI:10.1039/D3CP01388J