
A) Motif prediction analysis of the 68 TRF binding sites, performed... | Download Scientific Diagram

De novo prediction of Smad1/5 binding motif. Total 170 peak regions... | Download Scientific Diagram
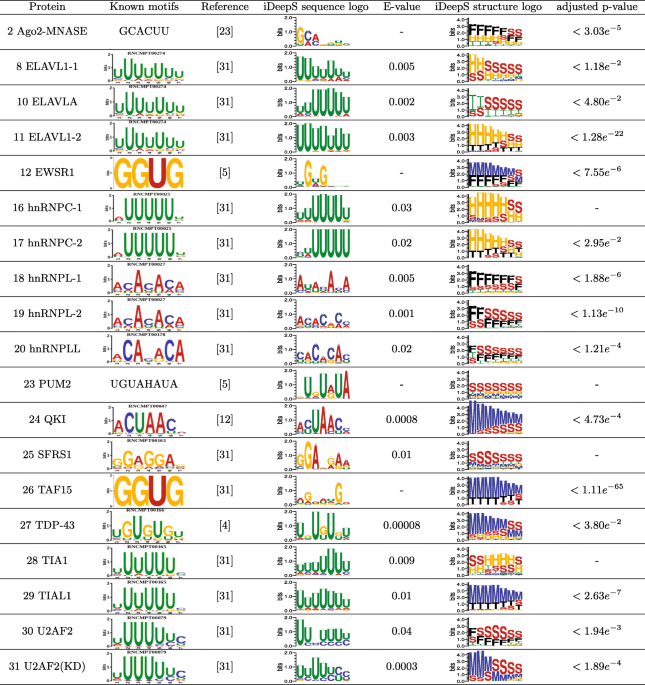
Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks | BMC Genomics | Full Text

PDF) Binding Site Prediction for Protein-Protein Interactions and Novel Motif Discovery using Re-occurring Polypeptide Sequences | James Green - Academia.edu
GitHub - morrislab/ATS-motif-prediction: Python scripts that predict RBP binding motifs based on target site accessibility in bound (positive) and unbound (negative) transcripts.
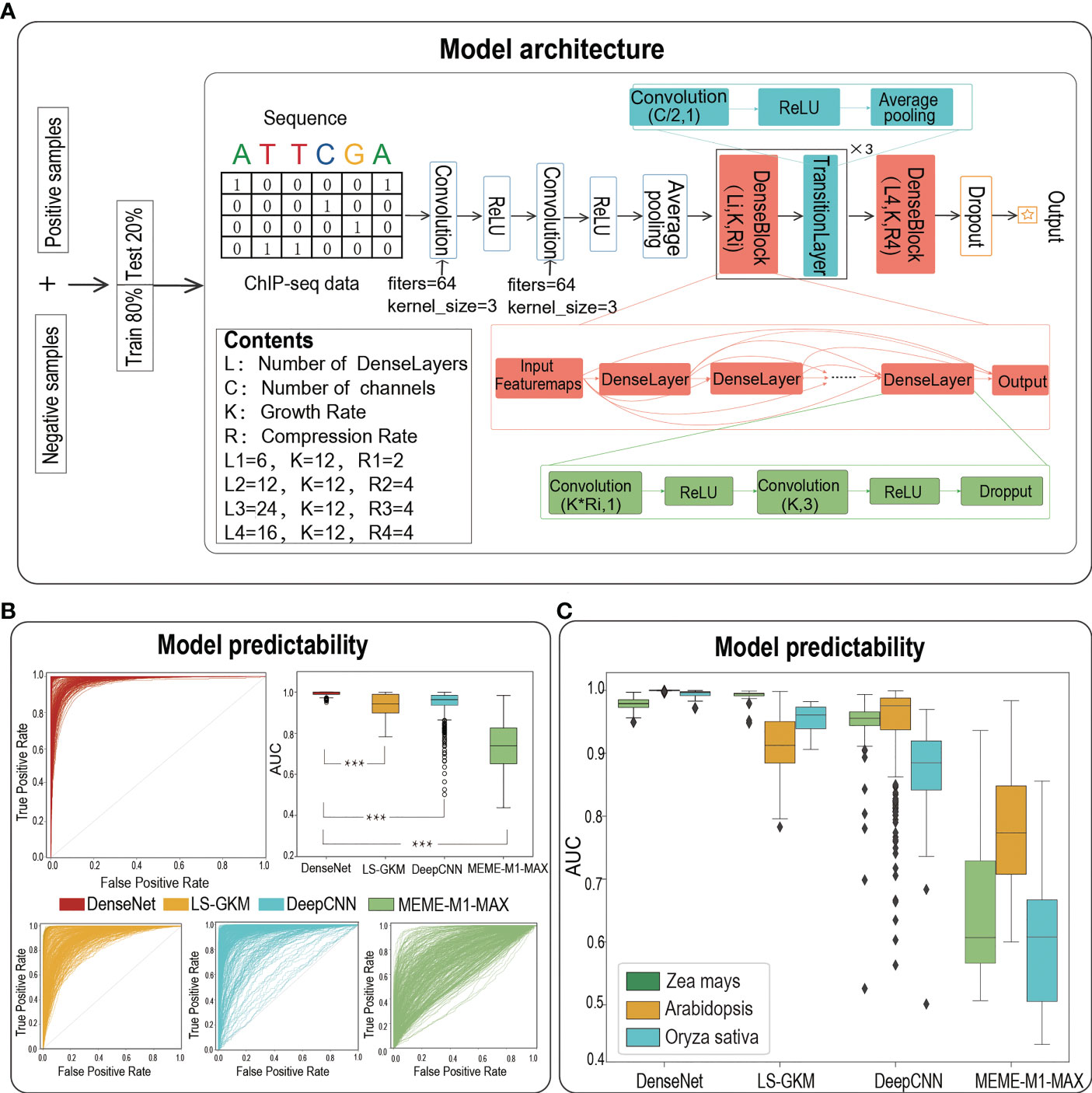
Frontiers | TSPTFBS 2.0: trans-species prediction of transcription factor binding sites and identification of their core motifs in plants
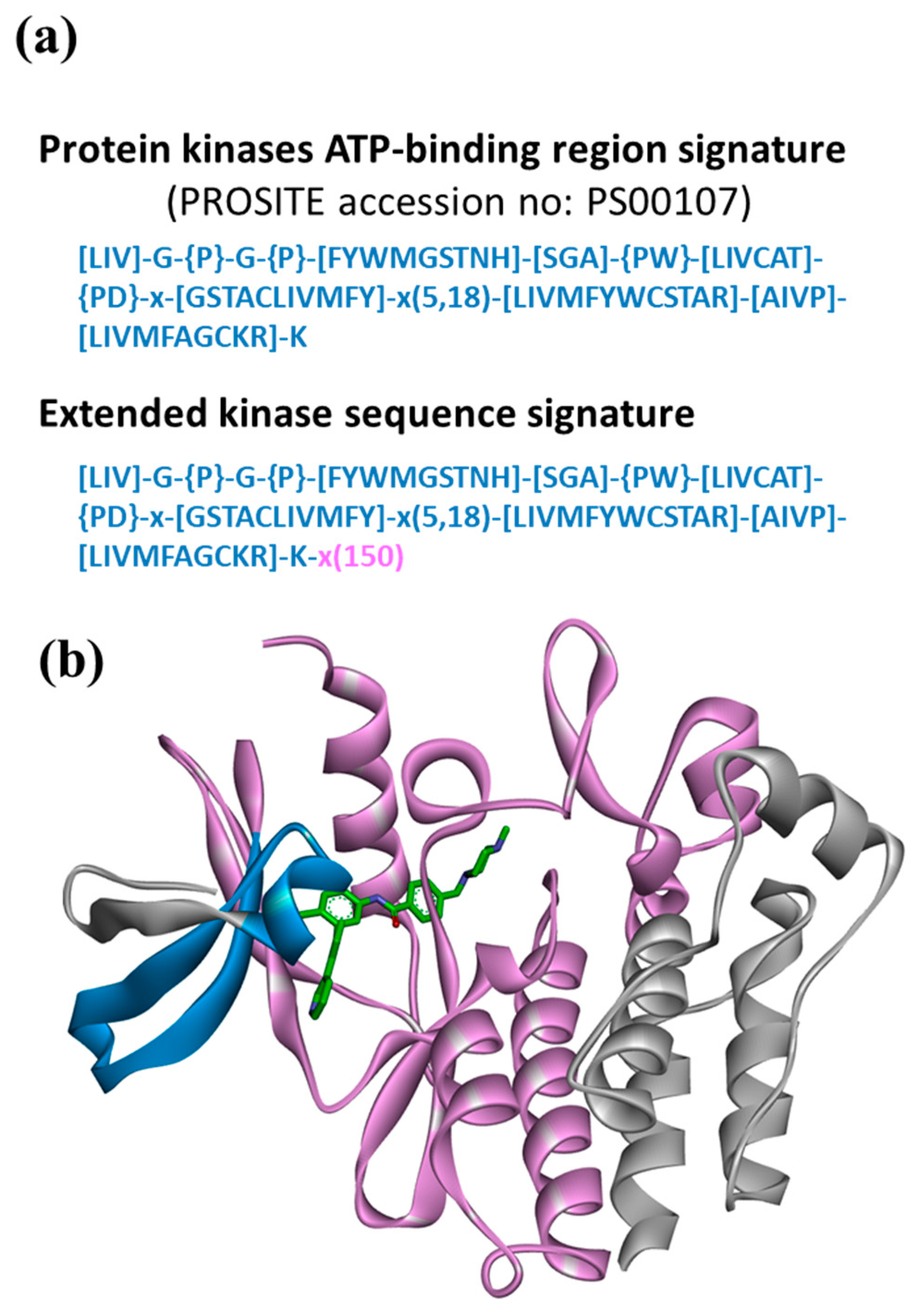
Biomolecules | Free Full-Text | Motif2Mol: Prediction of New Active Compounds Based on Sequence Motifs of Ligand Binding Sites in Proteins Using a Biochemical Language Model
![PDF] GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments | Semantic Scholar PDF] GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/30d1b6071a4b2c1434755cf4d8d7c0830cc07c14/2-Figure1-1.png)
PDF] GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments | Semantic Scholar
A Novel Bayesian DNA Motif Comparison Method for Clustering and Retrieval | PLOS Computational Biology

Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks | BMC Genomics | Full Text
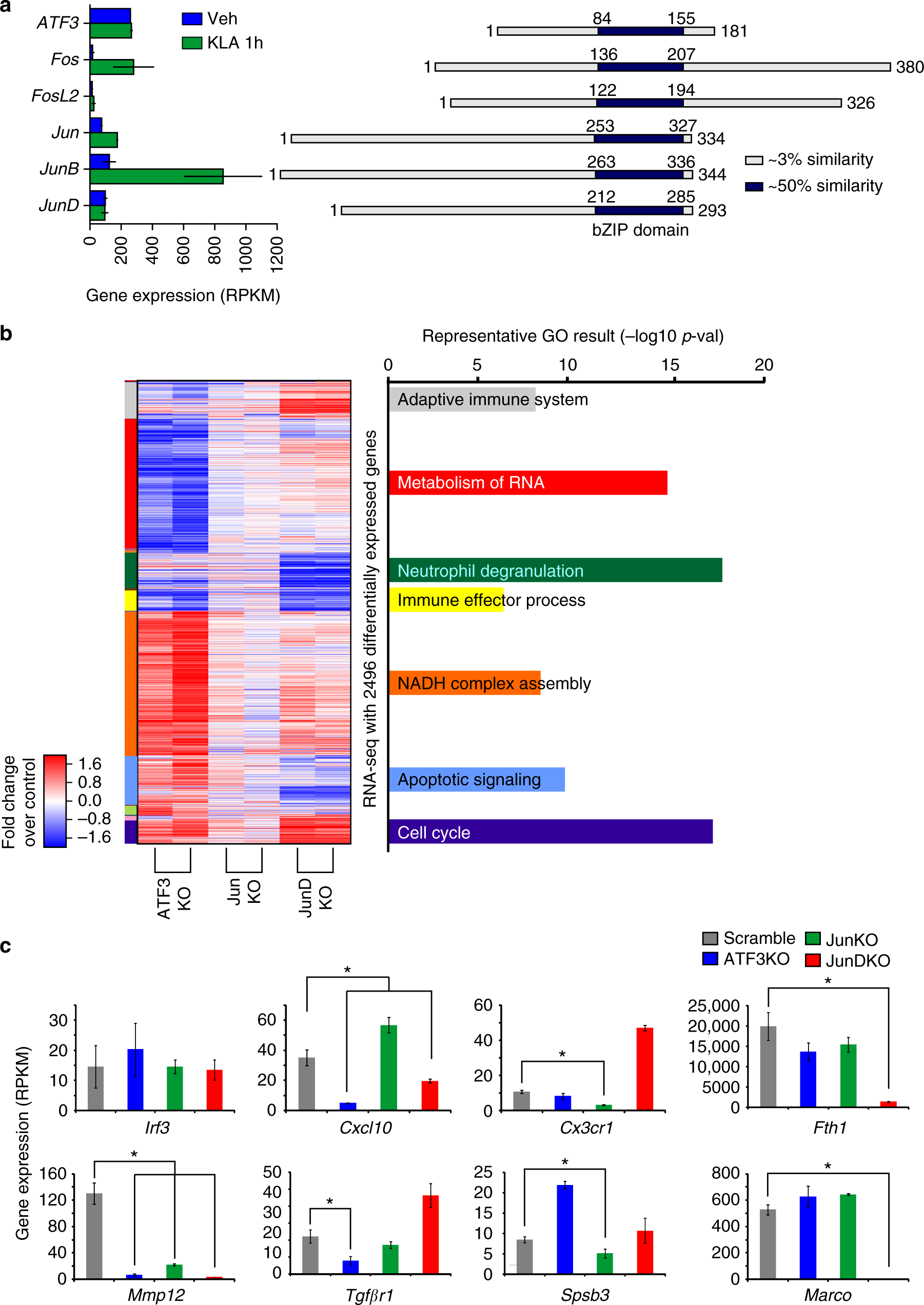
Diverse motif ensembles specify non-redundant DNA binding activities of AP-1 family members in macrophages | Nature Communications

Predicting the DNA Binding Site Motifs of Novel Transcription Factors... | Download Scientific Diagram

Prediction of DNA binding motifs from 3D models of transcription factors; identifying TLX3 regulated genes" published in Nucleic Acids Research — The Tapinos Lab
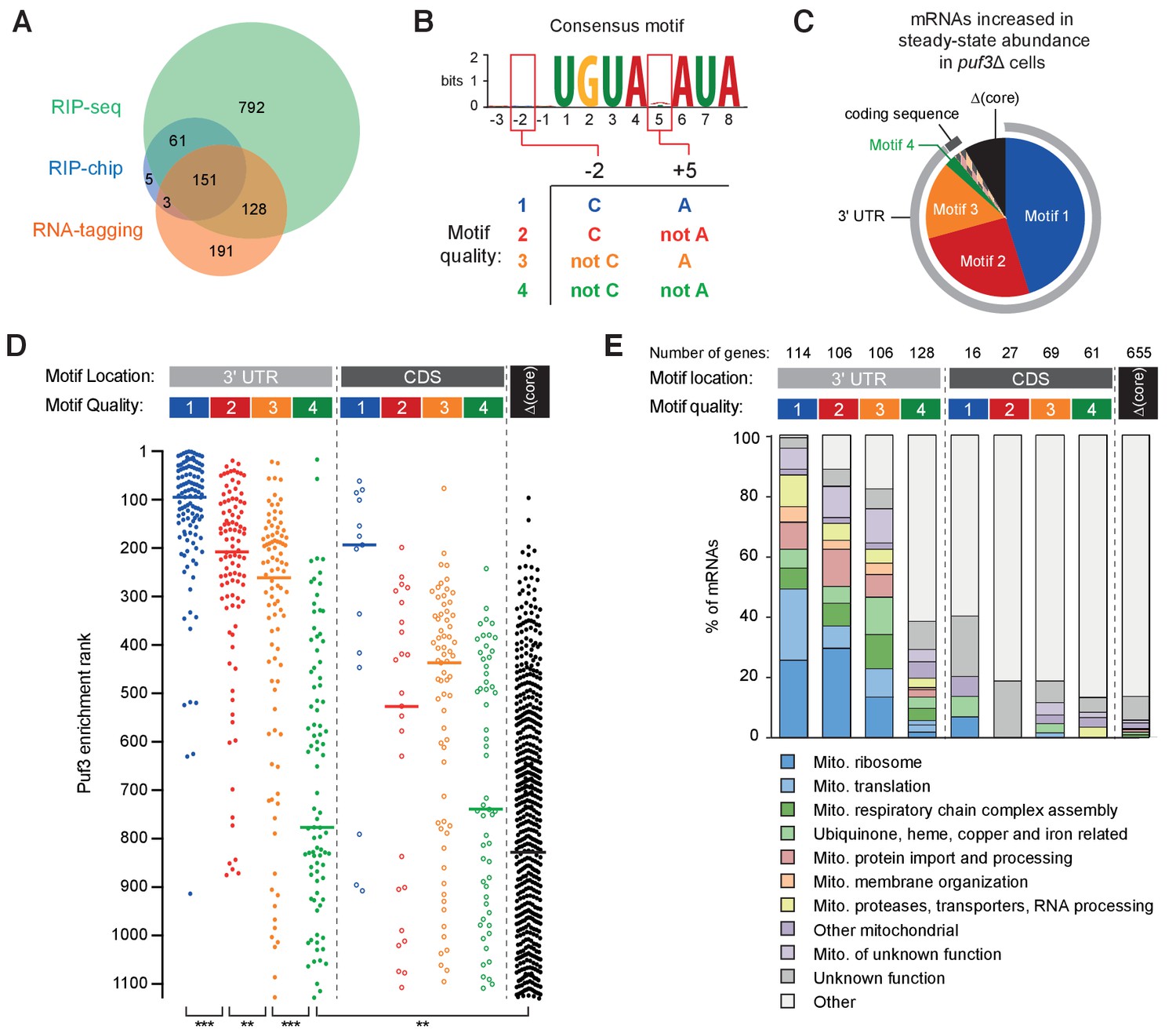
RNA-binding proteins distinguish between similar sequence motifs to promote targeted deadenylation by Ccr4-Not | eLife

De novo motif prediction on ESA1, GCN5 and SET1-binding sites. HOMER... | Download Scientific Diagram

Prediction of transcription factor binding sites and mutation analysis... | Download Scientific Diagram

Predicting transcription factor binding sites using DNA shape features based on shared hybrid deep learning architecture: Molecular Therapy - Nucleic Acids



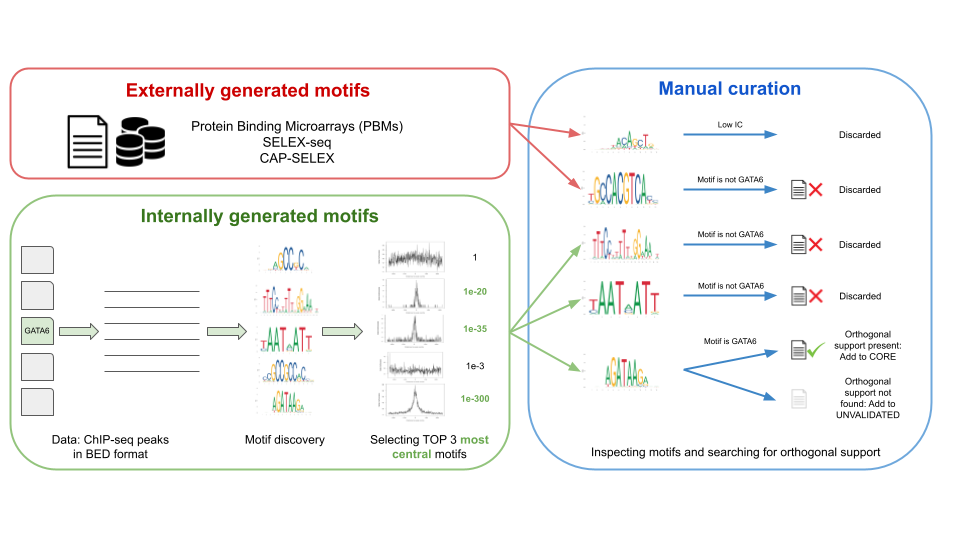

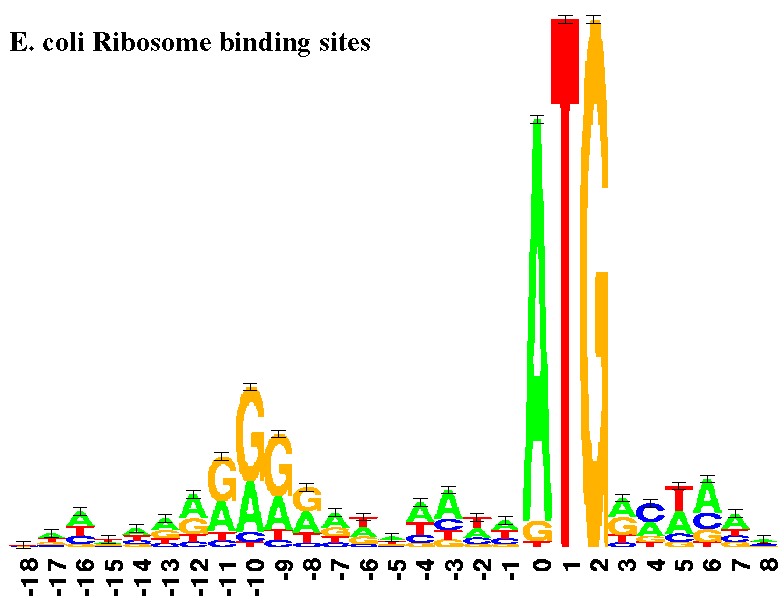
![PDF] Learning Methods for DNA Binding in Computational Biology | Semantic Scholar PDF] Learning Methods for DNA Binding in Computational Biology | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/6f6e44a74e2236e6840c9f9f7dbd225964fe5db1/58-Figure3-1.png)