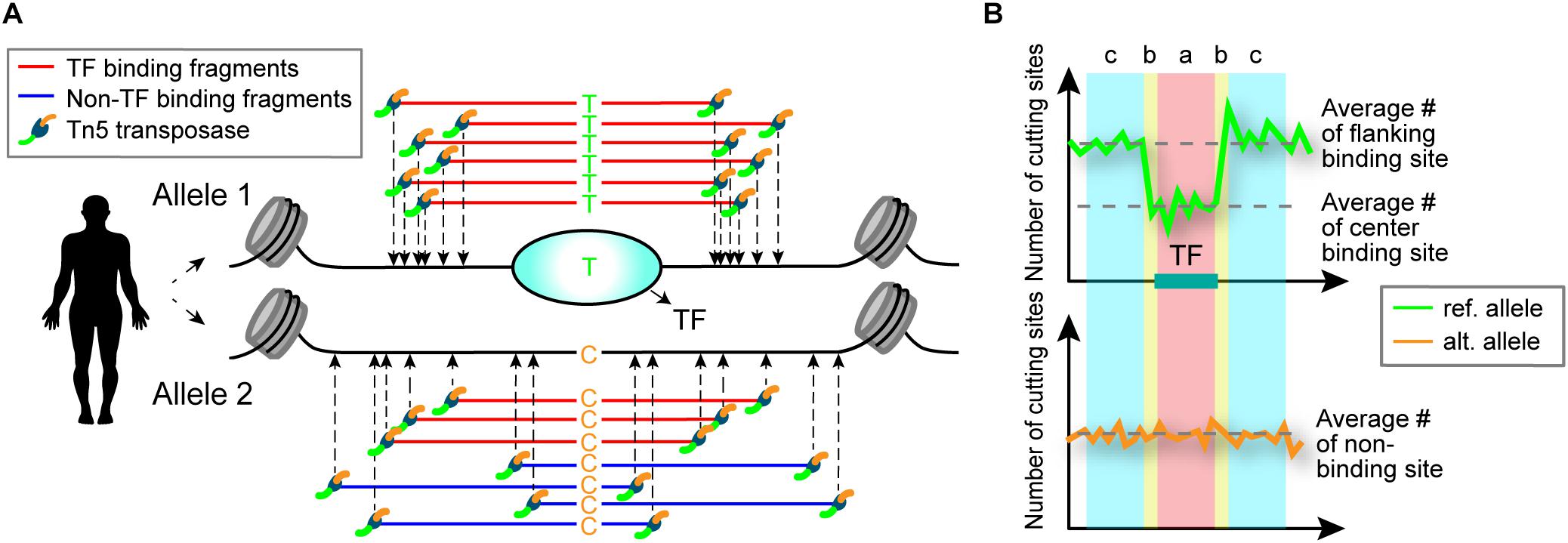
Frontiers | regSNPs-ASB: A Computational Framework for Identifying Allele-Specific Transcription Factor Binding From ATAC-seq Data
New Tool Provides Low-Cost, High-Quality Means to Map Transcription Factor Binding Genome-Wide - Research Horizons

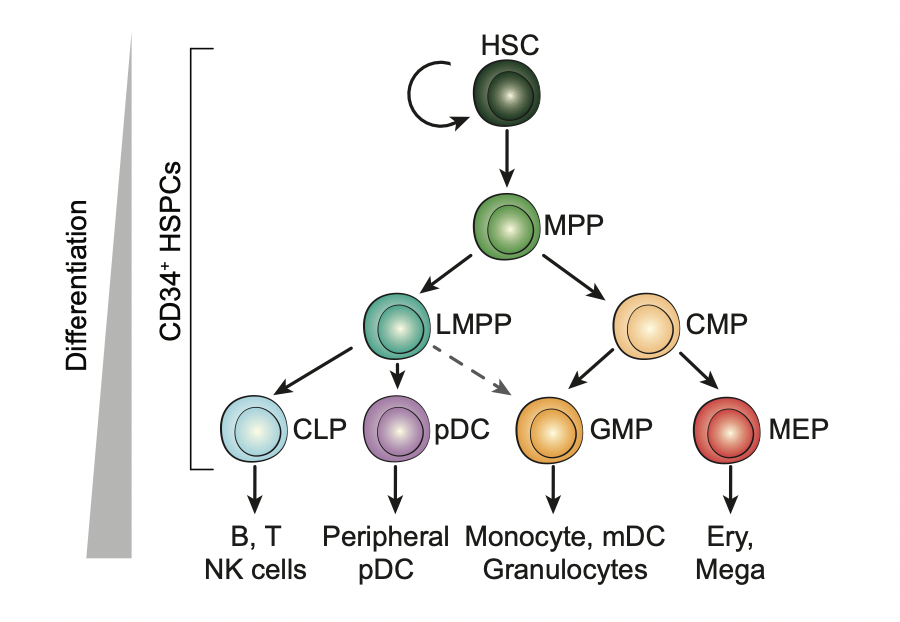
ATAC-Seq Reveals an Isl1 Enhancer That Regulates Sinoatrial Node Development and Function | Circulation Research

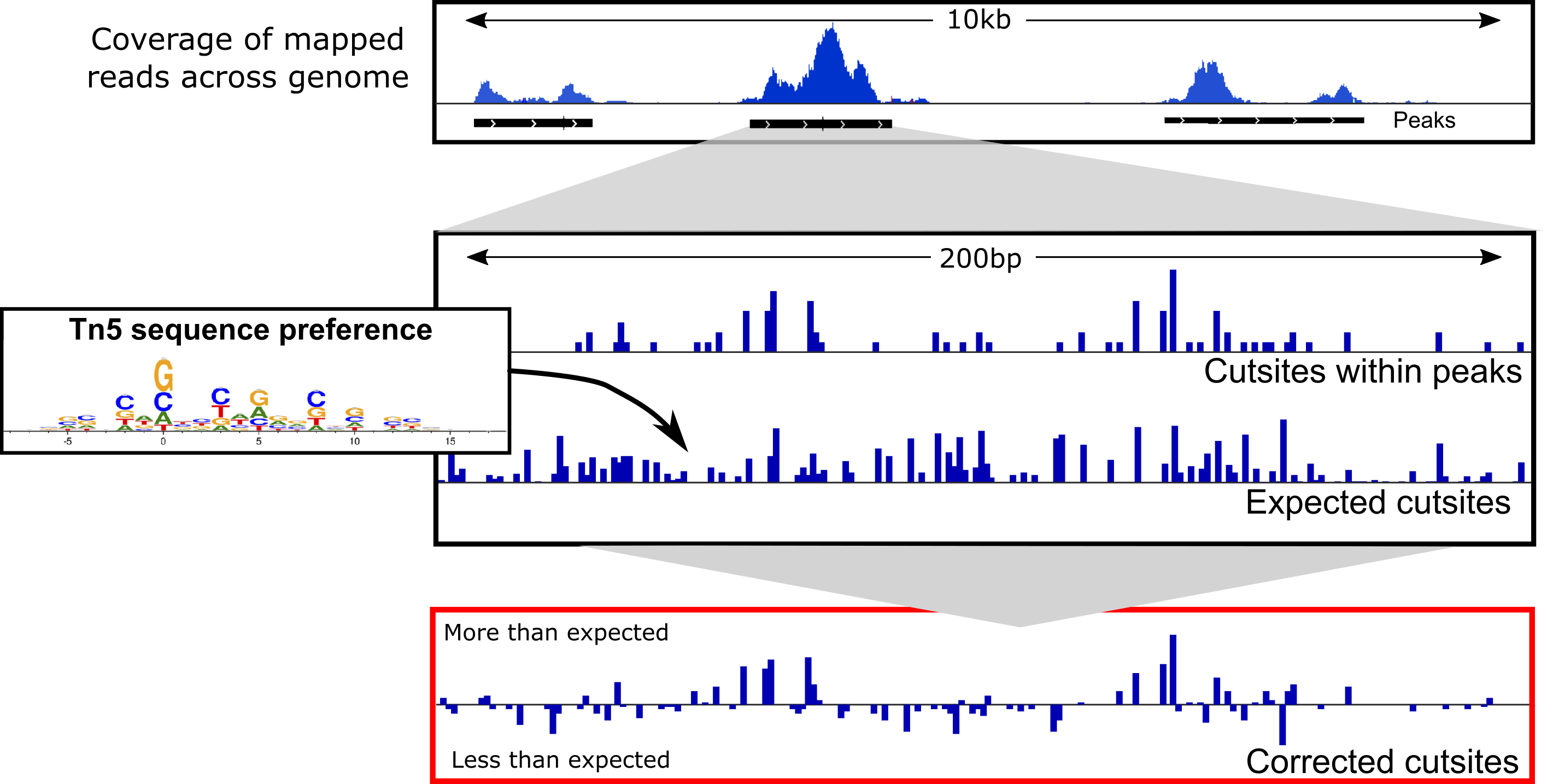
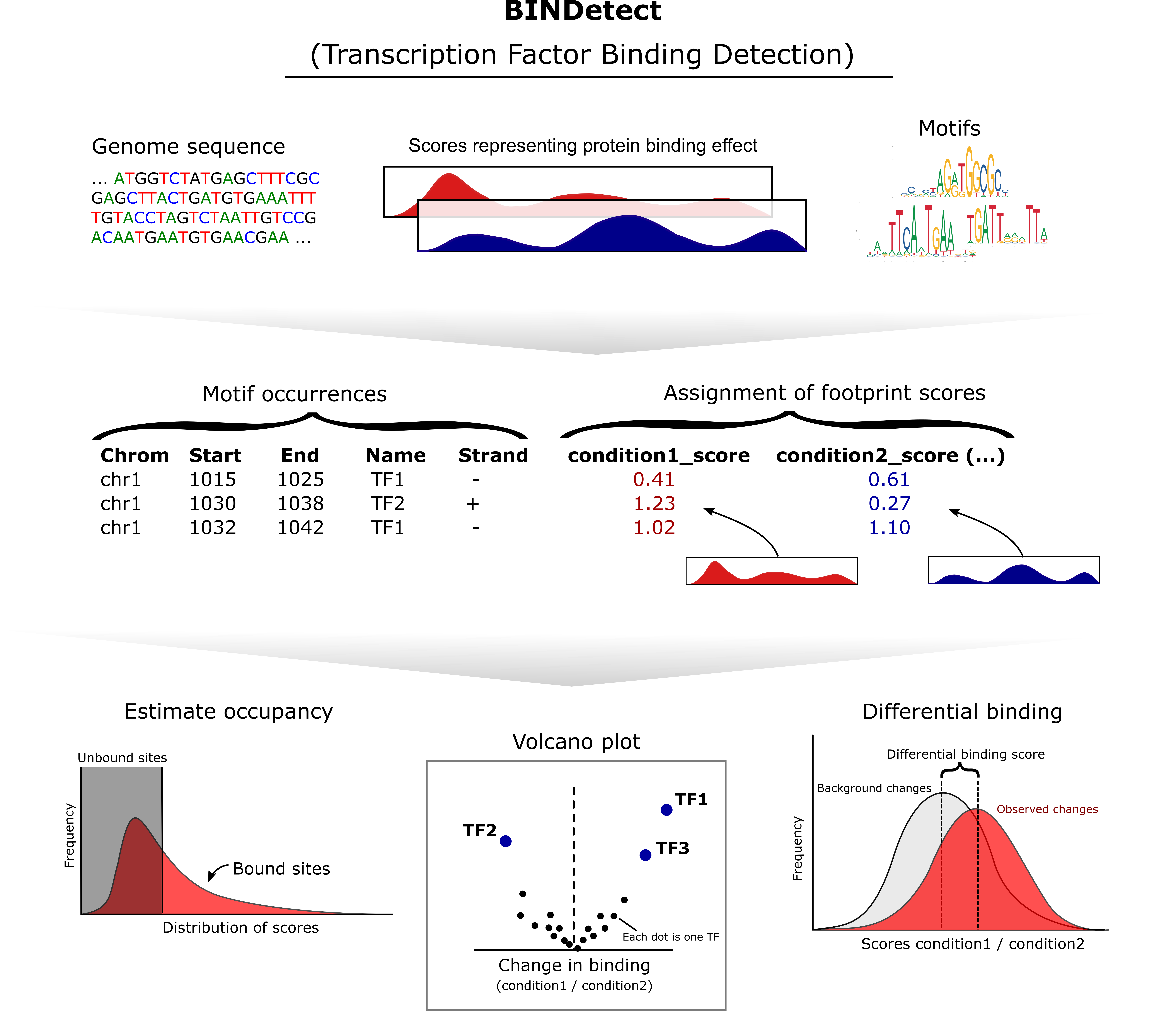
Beyond accessibility: ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation | bioRxiv
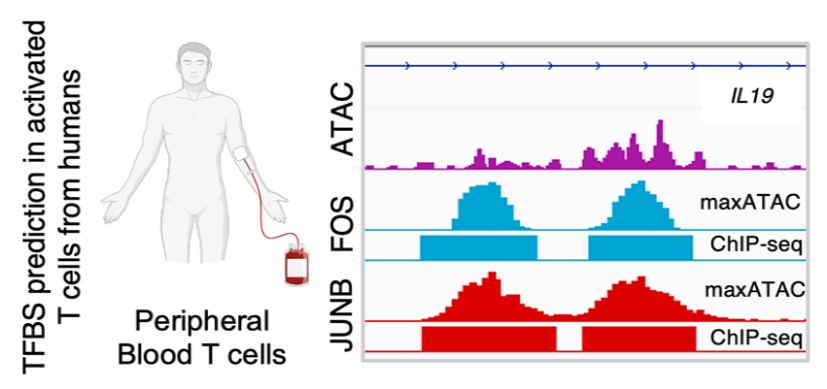
maxATAC: Genome-scale transcription-factor binding prediction from ATAC-seq with deep neural networks | PLOS Computational Biology

ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation | Nature Communications

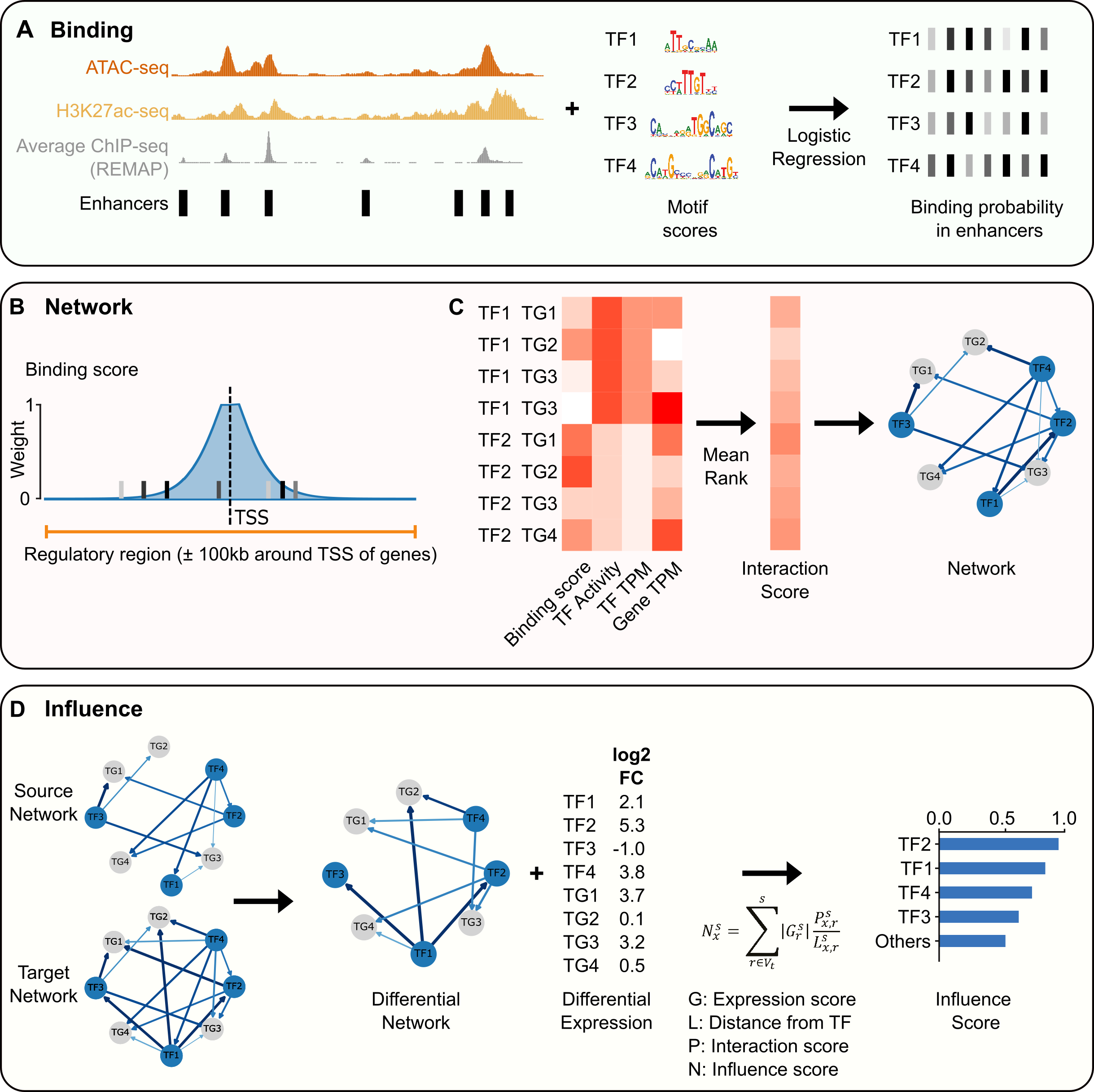
Quantification of absolute transcription factor binding affinities in the native chromatin context using BANC-seq | Nature Biotechnology

Transcription factor binding sites that are enriched in ATAC-Seq peaks... | Download Scientific Diagram

ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation | Nature Communications

Beyond accessibility: ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation | bioRxiv

ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation | Nature Communications